2025-05-05 ミュンヘン大学(LMU)
<関連情報>
- https://www.lmu.de/en/newsroom/news-overview/news/biological-patterns-stability-through-protein-reservoirs.html
- https://www.nature.com/articles/s41567-025-02878-w
生体内におけるMinタンパク質のロバストかつ資源最適な動的パターン形成 Robust and resource-optimal dynamic pattern formation of Min proteins in vivo
Ziyuan Ren,Henrik Weyer,Michael Sandler,Laeschkir Würthner,Haochen Fu,Chanin B. Tangtartharakul,Dongyang Li,Cindy Sou,Daniel Villarreal,Judy E. Kim,Erwin Frey & Suckjoon Jun
Nature Physics Published:05 May 2025
DOI:https://doi.org/10.1038/s41567-025-02878-w
Abstract
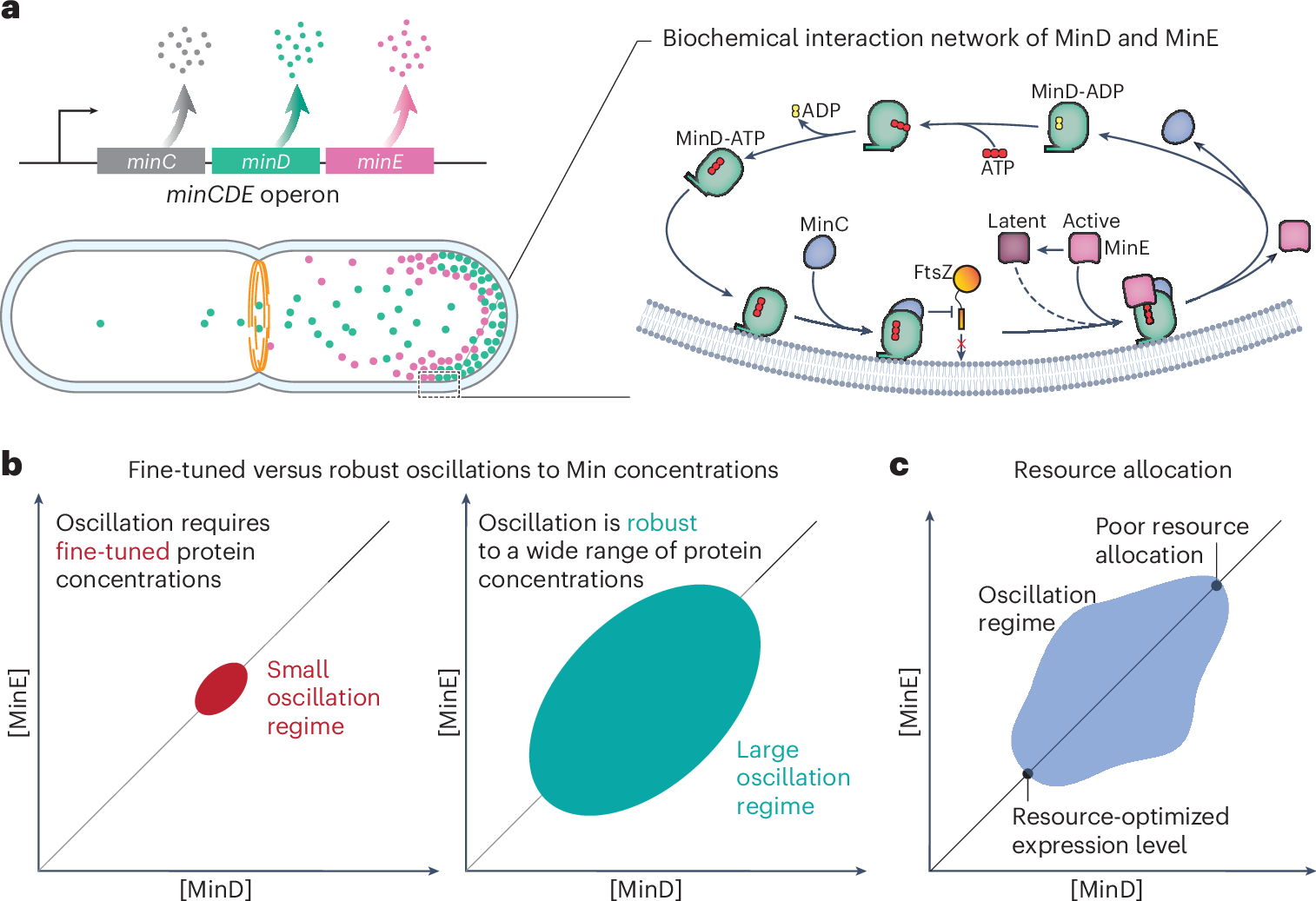
The Min protein system prevents abnormal cell division in bacteria by forming oscillatory patterns between cell poles. However, predicting the protein concentrations at which oscillations start and whether cells can maintain them under physiological perturbations remains challenging. Here we show that dynamic pattern formation is robust across a wide range of Min protein levels and variations in the growth physiology using genetically engineered Escherichia coli strains. We modulate the expression of minCD and minE under fast- and slow-growth conditions and build a MinD versus MinE phase diagram that reveals dynamic patterns, including travelling and standing waves. We found that the natural expression level of Min proteins is resource-optimal and robust to changes in protein concentration. In addition, we observed an invariant wavelength of dynamic Min patterns across the phase diagram. We explain the experimental findings quantitatively with biophysical theory based on reaction–diffusion models that consider the switching of MinE between its latent and active states, indicating its essential role as a robustness module for Min oscillation in vivo. Our results underline the potential of integrating quantitative cell physiology and biophysical modelling to understand the fundamental mechanisms controlling cell division machinery, and they offer insights applicable to other biological processes.