2025-07-09 マウントサイナイ医療システム (MSHS)
Credit: Campanella, et al., Nature Medicine
<関連情報>
- https://www.mountsinai.org/about/newsroom/2025/study-shows-how-ai-could-help-pathologists-match-cancer-patients-to-the-right-treatments-faster-and-more-efficiently
- https://www.nature.com/articles/s41591-025-03780-x
肺がんバイオマーカー検出のためのファインチューニング病理基盤モデルの実世界展開 Real-world deployment of a fine-tuned pathology foundation model for lung cancer biomarker detection
Gabriele Campanella,Neeraj Kumar,Swaraj Nanda,Siddharth Singi,Eugene Fluder,Ricky Kwan,Silke Muehlstedt,Nicole Pfarr,Peter J. Schüffler,Ida Häggström,Noora Neittaanmäki,Levent M. Akyürek,Alina Basnet,Tamara Jamaspishvili,Michel R. Nasr,Matthew M. Croken,Fred R. Hirsch,Arielle Elkrief,Helena Yu,Orly Ardon,Gregory M. Goldgof,Meera Hameed,Jane Houldsworth,Maria Arcila,… Chad Vanderbilt
Nature Medicine Published:09 July 2025
DOI:https://doi.org/10.1038/s41591-025-03780-x
Abstract
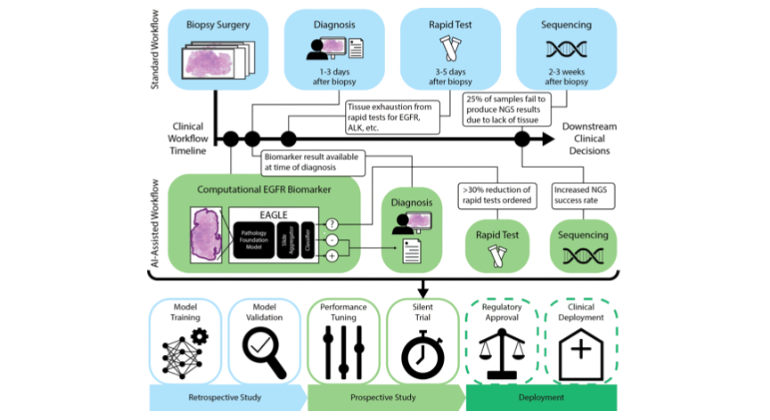
Artificial intelligence models using digital histopathology slides stained with hematoxylin and eosin offer promising, tissue-preserving diagnostic tools for patients with cancer. Despite their advantages, their clinical utility in real-world settings remains unproven. Assessing EGFR mutations in lung adenocarcinoma demands rapid, accurate and cost-effective tests that preserve tissue for genomic sequencing. PCR-based assays provide rapid results but with reduced accuracy compared with next-generation sequencing and require additional tissue. Computational biomarkers leveraging modern foundation models can address these limitations. Here we assembled a large international clinical dataset of digital lung adenocarcinoma slides (N = 8,461) to develop a computational EGFR biomarker. Our model fine-tunes an open-source foundation model, improving task-specific performance with out-of-center generalization and clinical-grade accuracy on primary and metastatic specimens (mean area under the curve: internal 0.847, external 0.870). To evaluate real-world clinical translation, we conducted a prospective silent trial of the biomarker on primary samples, achieving an area under the curve of 0.890. The artificial-intelligence-assisted workflow reduced the number of rapid molecular tests needed by up to 43% while maintaining the current clinical standard performance. Our retrospective and prospective analyses demonstrate the real-world clinical utility of a computational pathology biomarker.