2025-07-14 中国科学院(CAS)
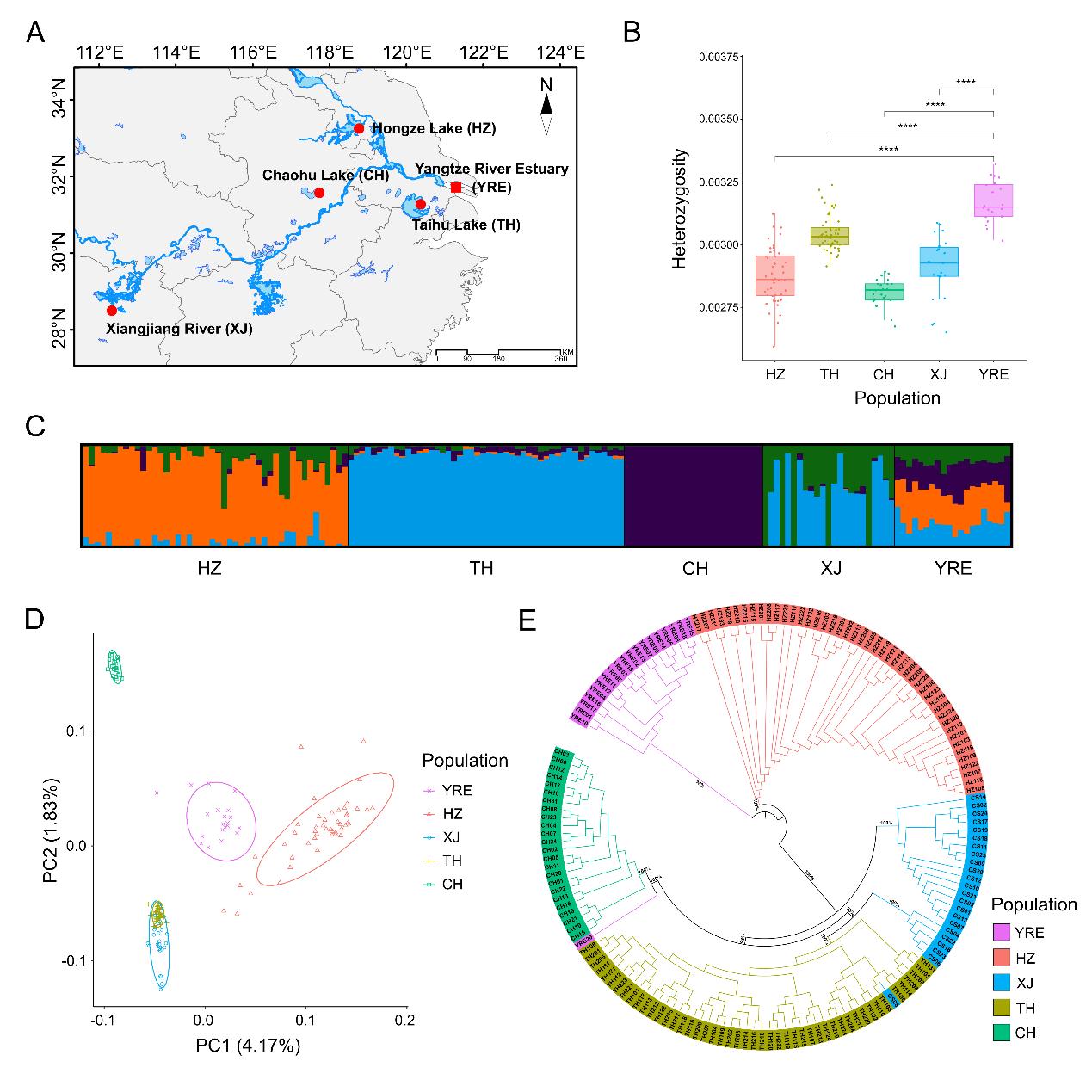 Sampling map, genetic diversity, and population genetic structure of Neosalanx brevirostris populations. (Image by IOCAS)
Sampling map, genetic diversity, and population genetic structure of Neosalanx brevirostris populations. (Image by IOCAS)
<関連情報>
- https://english.cas.cn/newsroom/research_news/earth/202507/t20250714_1047332.shtml
- https://academic.oup.com/mbe/advance-article/doi/10.1093/molbev/msaf160/8185571
野生魚の常在遺伝子変異がもたらす淡水への急速な適応を支えるゲノムワイドな並列性 Genome-wide Parallelism Underlies Rapid Freshwater Adaptation Fueled by Standing Genetic Variation in a Wild Fish
Hao Yang , Yu-Long Li , Teng-Fei Xing , Jian-Hui Wu , Ting Wang , Ming-Sheng Zhu , Jin-Xian Liu
Molecular Biology and Evolution Published:03 July 2025
DOI:https://doi.org/10.1093/molbev/msaf160
Abstract
A fundamental focus of ecological and evolutionary biology is determining how natural populations adapt to environmental changes. Rapid parallel phenotypic evolution can be leveraged to uncover the genetics of adaptation. Using population genomic approaches, we investigated the genetic architecture underlying rapid parallel freshwater adaptation of Neosalanx brevirostris by comparing four freshwater-resident populations with their common ancestral anadromous population. We demonstrated that the rapid parallel adaptation to freshwater followed a complex polygenic architecture and was characterised by genomic-level parallelism, which proceeded predominantly through repeated selection on the pre-existing standing genetic variations. Frequencies of the genome-wide adaptive standing variations were moderate in the ancestral anadromous population, which had pre-adapted to fluctuating salinities. Relatively large allele frequency shifts were observed at some adaptive SNPs during parallel adaptation to freshwater environments, with a large fraction of freshwater favored alleles being fixed or nearly fixed. These adaptive SNPs were involved in multiple biological functions associated with osmoregulation, immunoregulation, locomotion, metabolism, etc., which were highly consistent with the polygenic architecture of adaptive divergence between the two ecotypes involving multiple complex physiological and behavioral traits. This work provides insight into the mechanisms by which natural populations rapidly evolve to changes in the environment and highlights the importance of standing genetic variation for evolutionary potential of populations facing global environmental changes.