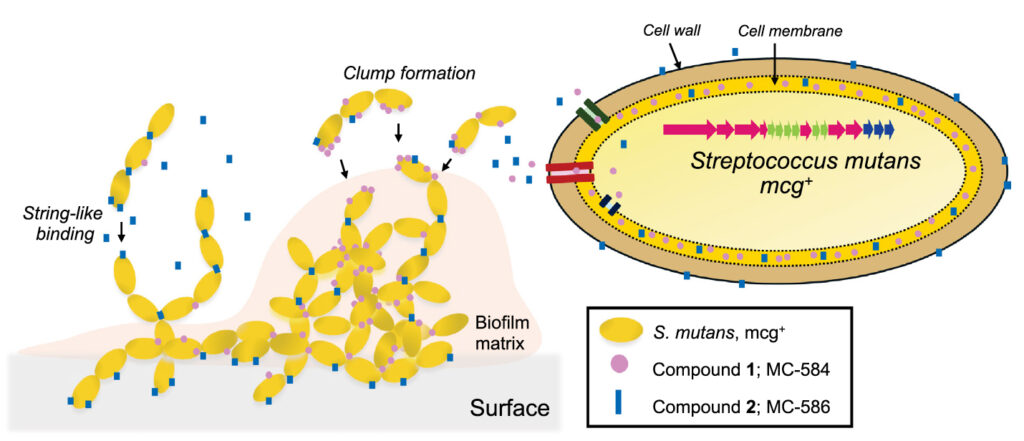
2025-09-04 パデュー大学
<関連情報>
- https://www.purdue.edu/newsroom/2025/Q3/blueprints-for-a-molecular-machine-more-powerful-than-crispr/?utm_source=chatgpt.com
- https://www.cell.com/cell/abstract/S0092-8674(24)01071-7
- https://www.cell.com/cell/fulltext/S0092-8674(23)00743-2
TnsABCDトランスポゾソームの構造解析が解明する標的DNA転移のメカニズム Structure of TnsABCD transpososome reveals mechanisms of targeted DNA transposition
Shukun Wang ∙ Romana Siddique ∙ Mark C. Hall ∙ Phoebe A. Rice ∙ Leifu Chang
Cell Published:October 8, 2024
DOI:https://doi.org/10.1016/j.cell.2024.09.023
Graphical abstract

Highlights
- Cryo-EM structure of the TnsC-TnsD-att DNA shows DNA bending featured by intercalation
- TnsABCD transpososome structure reveals TnsB recruitment to TnsC via TnsB’s C-tail
- TnsC’s C-tail directly participates in TnsAB-DNA strand-transfer complex assembly
- Structural details of the active sites of TnsA and TnsB within the transpososome
Summary
Tn7-like transposons are characterized by their ability to insert specifically into host chromosomes. Recognition of the attachment (att) site by TnsD recruits the TnsABC proteins to form the transpososome and facilitate transposition. Although this pathway is well established, atomic-level structural insights of this process remain largely elusive. Here, we present the cryo-electron microscopy (cryo-EM) structures of the TnsC-TnsD-att DNA complex and the TnsABCD transpososome from the Tn7-like transposon in Peltigera membranacea cyanobiont 210A, a type I-B CRISPR-associated transposon. Our structures reveal a striking bending of the att DNA, featured by the intercalation of an arginine side chain of TnsD into a CC/GG dinucleotide step. The TnsABCD transpososome structure reveals TnsA-TnsB interactions and demonstrates that TnsC not only recruits TnsAB but also directly participates in the transpososome assembly. These findings provide mechanistic insights into targeted DNA insertion by Tn7-like transposons, with implications for improving the precision and efficiency of their genome-editing applications.
I-B型CRISPRエフェクターによるTn7様トランスポゾン動員分子機構 Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector
Shukun Wang ∙ Clinton Gabel ∙ Romana Siddique ∙ Thomas Klose ∙ Leifu Chang
Cell Published:August 8, 2023
DOI:https://doi.org/10.1016/j.cell.2023.07.010
Highlights
- Biochemical reconstitution and discovery of Cas11 in type I-B2 PmcCAST
- Cryo-EM structure of the Cascade-DNA-TniQ-TnsC recruitment complex
- Target DNA binding promotes conformational changes in Cascade to recruit TniQ
- TniQ binds to the seam region of the TnsC spiral heptamer
Summary
Tn7-like transposons have co-opted CRISPR-Cas systems to facilitate the movement of their own DNA. These CRISPR-associated transposons (CASTs) are promising tools for programmable gene knockin. A key feature of CASTs is their ability to recruit Tn7-like transposons to nuclease-deficient CRISPR effectors. However, how Tn7-like transposons are recruited by diverse CRISPR effectors remains poorly understood. Here, we present the cryo-EM structure of a recruitment complex comprising the Cascade complex, TniQ, TnsC, and the target DNA in the type I-B CAST from Peltigera membranacea cyanobiont 210A. Target DNA recognition by Cascade induces conformational changes in Cas6 and primes TniQ recruitment through its C-terminal domain. The N-terminal domain of TniQ is bound to the seam region of the TnsC spiral heptamer. Our findings provide insights into the diverse mechanisms for the recruitment of Tn7-like transposons to CRISPR effectors and will aid in the development of CASTs as gene knockin tools.