2025-09-05 産業技術総合研究所
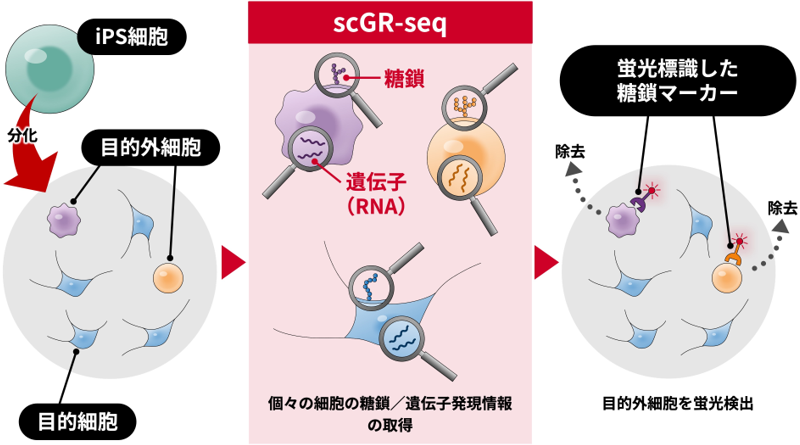
scGR-seq法によるiPS細胞由来分化細胞の糖鎖マーカー開発
<関連情報>
- https://www.aist.go.jp/aist_j/press_release/pr2025/pr20250905/pr20250905.html
- https://www.cell.com/stem-cell-reports/fulltext/S2213-6711(25)00235-8
単一細胞グリコミクスおよびトランスクリプトームプロファイリングにより、ヒト iPSC 由来ニューロンの各細胞サブポピュレーションの糖鎖シグネチャが明らかになる Single-cell glycome and transcriptome profiling uncovers the glycan signature of each cell subpopulation of human iPSC-derived neurons
Haruki Odaka ∙ Hiroaki Tateno
Stem Cell Reports September: 4, 2025
DOI:https://doi.org/10.1016/j.stemcr.2025.102631
Highlights
- Single-cell glycan and RNA sequencing revealed heterogeneity in iPSC-derived neurons
- Undifferentiated NPCs and mesenchymal cells (MCs) were identified as non-neural cells
- Glycan markers of undifferentiated NPCs and MCs were identified
- Glycan markers enabled the detection of non-neural cells in neural cultures
Summary
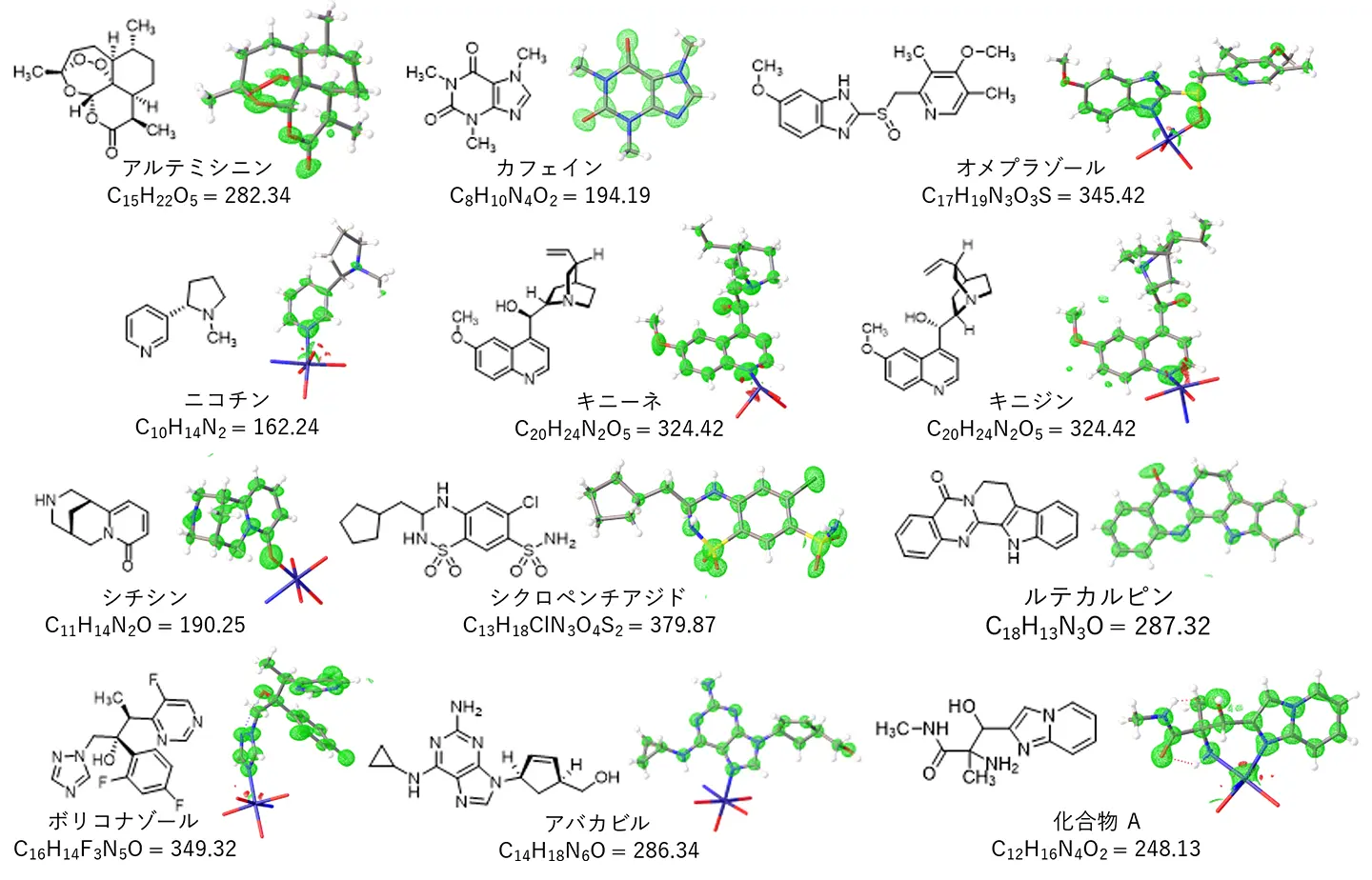
Human induced pluripotent stem cell (iPSC)-derived neurons are often heterogeneous, posing challenges for disease modeling and cell therapy. We previously developed single-cell glycan and RNA sequencing (scGR-seq) to analyze the glycome and transcriptome simultaneously. Here, we applied scGR-seq to examine heterogeneous populations of human iPSC-derived neurons. We identified four subpopulations: mature neurons, immature neurons, undifferentiated neural progenitor cells (undiffNPCs), and mesenchymal cells (MCs). Lectin-binding patterns indicated high α1,3-fucose expression in undiffNPCs. MCs exhibited strong binding of a poly-LacNAc-recognizing lectin (rLSLN) and high expression of B3GNT2, a poly-LacNAc synthetic enzyme. Pseudotime analysis revealed that a subpopulation of NPCs acquired mesenchymal features and differentiated into MCs. Immunocytochemistry confirmed the specific detection of undiffNPCs and MCs using anti-Lewis X (α1,3-fucosylated glycan) antibodies and rLSLN. Beyond identifying cell heterogeneity, scGR-seq enables the discovery of glycan markers and detection probes for iPSC-derived cells, aiding in their further cell processing and manipulation.