2022-09-06 オークリッジ国立研究所(ORNL)
研究チームは13,000以上のProtein Data Bank構造のトポロジーを分析し、ウイルスのスパイクタンパク質の突然変異は、タンパク質が安定でなくなるトポロジカル自由エネルギーの高い領域で最も発生しやすいことを発見した。
SARS-CoV-2 omicron変異体は、研究者の計算と一致する領域で変異している。
この方法は、タンパク質の変異がどこで起こるかを数学的に予測するもので、あらゆるタンパク質に適用可能であり、ウイルスの動態の理解やより効率的な薬物治療への扉を開く可能性がある。
<関連情報>
- https://www.ornl.gov/news/using-math-predict-sars-cov-2-protein-mutations
- https://www.mdpi.com/2073-4360/14/15/3014/htm
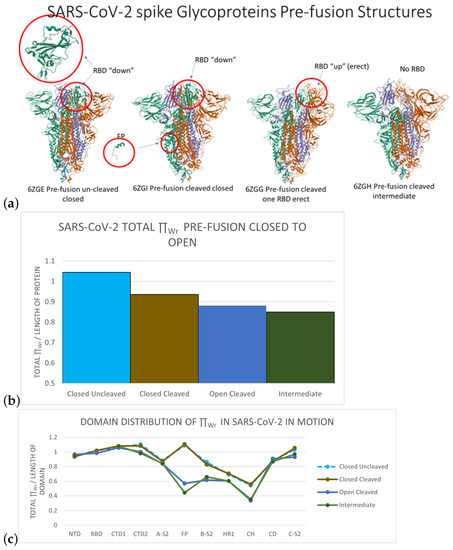
SARS-CoV-2スパイクタンパク質の局所トポロジカル自由エネルギー The Local Topological Free Energy of the SARS-CoV-2 Spike Protein
Quenisha Baldwin,Bobby Sumpter and Eleni Panagiotou
Polymers Published: 26 July 2022
DOI:https://doi.org/10.3390/polym14153014
Abstract
The novel coronavirus SARS-CoV-2 infects human cells using a mechanism that involves binding and structural rearrangement of its Spike protein. Understanding protein rearrangement and identifying specific amino acids where mutations affect protein rearrangement has attracted much attention for drug development. In this manuscript, we use a mathematical method to characterize the local topology/geometry of the SARS-CoV-2 Spike protein backbone. Our results show that local conformational changes in the FP, HR1, and CH domains are associated with global conformational changes in the RBD domain. The SARS-CoV-2 variants analyzed in this manuscript (alpha, beta, gamma, delta Mink, G614, N501) show differences in the local conformations of the FP, HR1, and CH domains as well. Finally, most mutations of concern are either in or in the vicinity of high local topological free energy conformations, suggesting that high local topological free energy conformations could be targets for mutations with significant impact of protein function. Namely, the residues 484, 570, 614, 796, and 969, which are present in variants of concern and are targeted as important in protein function, are predicted as such from our model.