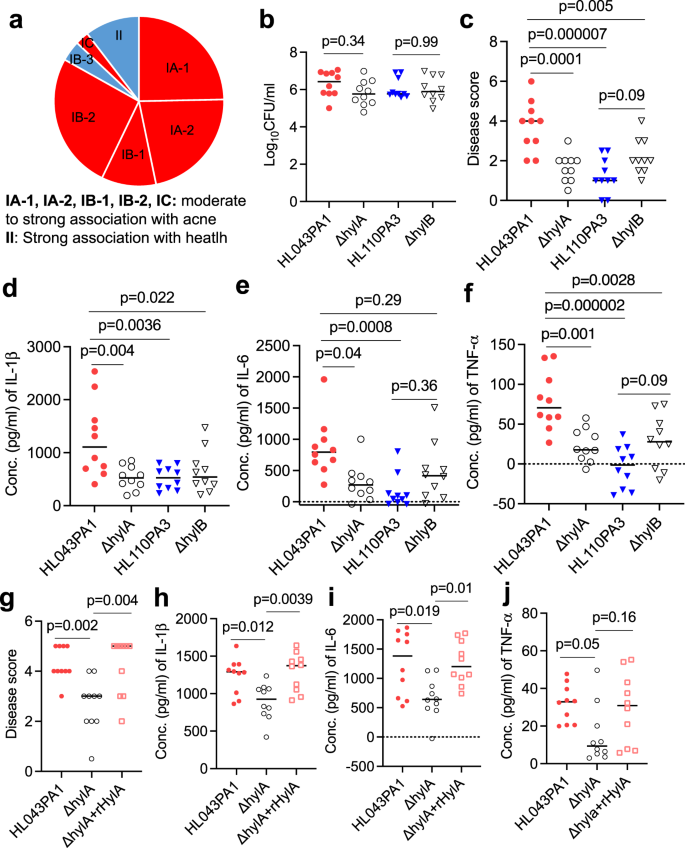
2023-12-20 ジョージア工科大学
◆これにより、タンパク質や代謝物などの詳細な情報を取得し、患者の生体物理学的なメイクアップを理解して最適な治療法を見つける可能性があります。Coskun氏は、この手法が将来的にはがんや女性の疾患、代謝性障害などの効果的な管理に貢献すると期待しています。
<関連情報>
- https://research.gatech.edu/coskun-lab-pioneering-new-field-research-single-cell-spatial-metabolomics
- https://www.nature.com/articles/s41467-023-43917-5#Ack1
- https://www.science.org/doi/10.1126/sciadv.abd0957
組織システム生物学のための細胞タイプ特異的タンパク質プロファイリングを用いたシングルセル空間メタボロミクス Single-cell spatial metabolomics with cell-type specific protein profiling for tissue systems biology
Thomas Hu,Mayar Allam,Shuangyi Cai,Walter Henderson,Brian Yueh,Aybuke Garipcan,Anton V. Ievlev,Maryam Afkarian,Semir Beyaz & Ahmet F. Coskun
Nature Communications Published:13 December 2023
DOI:https://doi.org/10.1038/s41467-023-43917-5
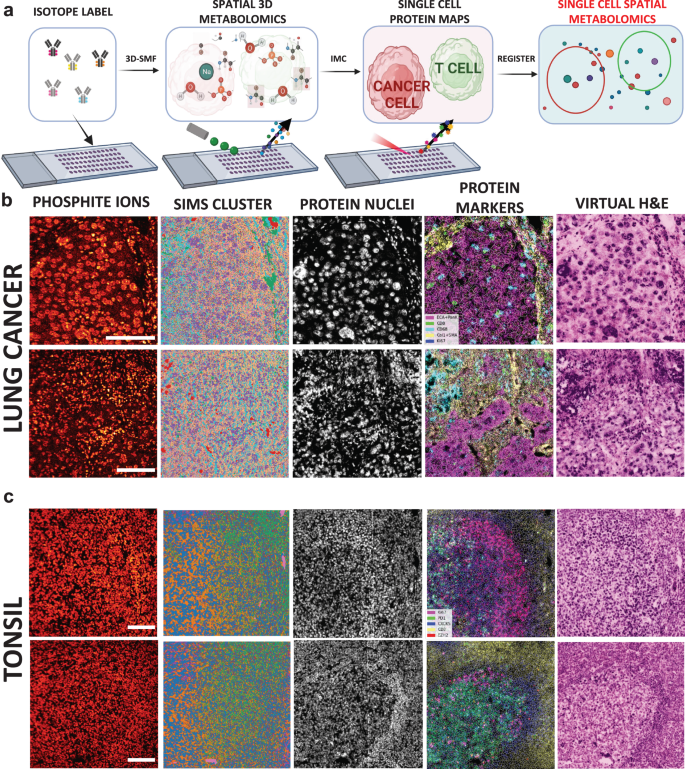
Abstract
Metabolic reprogramming in cancer and immune cells occurs to support their increasing energy needs in biological tissues. Here we propose Single Cell Spatially resolved Metabolic (scSpaMet) framework for joint protein-metabolite profiling of single immune and cancer cells in male human tissues by incorporating untargeted spatial metabolomics and targeted multiplexed protein imaging in a single pipeline. We utilized the scSpaMet to profile cell types and spatial metabolomic maps of 19507, 31156, and 8215 single cells in human lung cancer, tonsil, and endometrium tissues, respectively. The scSpaMet analysis revealed cell type-dependent metabolite profiles and local metabolite competition of neighboring single cells in human tissues. Deep learning-based joint embedding revealed unique metabolite states within cell types. Trajectory inference showed metabolic patterns along cell differentiation paths. Here we show scSpaMet’s ability to quantify and visualize the cell-type specific and spatially resolved metabolic-protein mapping as an emerging tool for systems-level understanding of tissue biology.
組織における空間分解3Dメタボロームプロファイリング Spatially resolved 3D metabolomic profiling in tissues
Shambavi Ganesh,Thomas Hu,Eric Woods,Mayar Allam,Shuangyi Cai,Walter Henderson,and Ahmet F. Coskun
Science Advances Published:27 Jan 2021
DOI:https://doi.org/10.1126/sciadv.abd0957

Abstract
Spatially resolved RNA and protein molecular analyses have revealed unexpected heterogeneity of cells. Metabolic analysis of individual cells complements these single-cell studies. Here, we present a three-dimensional spatially resolved metabolomic profiling framework (3D-SMF) to map out the spatial organization of metabolic fragments and protein signatures in immune cells of human tonsils. In this method, 3D metabolic profiles were acquired by time-of-flight secondary ion mass spectrometry to profile up to 189 compounds. Ion beams were used to measure sub–5-nanometer layers of tissue across 150 sections of a tonsil. To incorporate cell specificity, tonsil tissues were labeled by an isotope-tagged antibody library. To explore relations of metabolic and cellular features, we carried out data reduction, 3D spatial correlations and classifications, unsupervised K-means clustering, and network analyses. Immune cells exhibited spatially distinct lipidomic fragment distributions in lymphatic tissue. The 3D-SMF pipeline affects studying the immune cells in health and disease.