2024-04-08 プリンストン大学
<関連情報>
- https://engineering.princeton.edu/news/2024/04/08/can-language-models-read-genome-one-decoded-mrna-make-better-vaccines
- https://www.nature.com/articles/s42256-024-00823-9
mRNAの非翻訳領域の解読と機能予測のための5′UTR言語モデル A 5′ UTR language model for decoding untranslated regions of mRNA and function predictions
Yanyi Chu,Dan Yu,Yupeng Li,Kaixuan Huang,Yue Shen,Le Cong,Jason Zhang & Mengdi Wang
Nature Machine Intelligence Published:05 April 2024
DOI:https://doi.org/10.1038/s42256-024-00823-9
Abstract
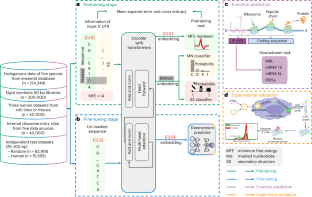
The 5′ untranslated region (UTR), a regulatory region at the beginning of a messenger RNA (mRNA) molecule, plays a crucial role in regulating the translation process and affects the protein expression level. Language models have showcased their effectiveness in decoding the functions of protein and genome sequences. Here, we introduce a language model for 5′ UTR, which we refer to as the UTR-LM. The UTR-LM is pretrained on endogenous 5′ UTRs from multiple species and is further augmented with supervised information including secondary structure and minimum free energy. We fine-tuned the UTR-LM in a variety of downstream tasks. The model outperformed the best known benchmark by up to 5% for predicting the mean ribosome loading, and by up to 8% for predicting the translation efficiency and the mRNA expression level. The model was also applied to identifying unannotated internal ribosome entry sites within the untranslated region and improved the area under the precision–recall curve from 0.37 to 0.52 compared to the best baseline. Further, we designed a library of 211 new 5′ UTRs with high predicted values of translation efficiency and evaluated them via a wet-laboratory assay. Experiment results confirmed that our top designs achieved a 32.5% increase in protein production level relative to well-established 5′ UTRs optimized for therapeutics.