2024-05-21 カリフォルニア大学サンディエゴ校(UCSD)
<関連情報>
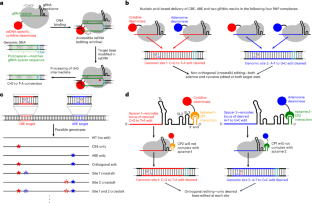
多重化直交ベースエディター(MOBE)システムの開発 Development of multiplexed orthogonal base editor (MOBE) systems
Quinn T. Cowan,Sifeng Gu,Wanjun Gu,Brodie L. Ranzau,Tatum S. Simonson & Alexis C. Komor
Nature Biotechnology Published:21 May 2024
DOI:https://doi.org/10.1038/s41587-024-02240-0
Abstract
Base editors (BEs) enable efficient, programmable installation of point mutations while avoiding the use of double-strand breaks. Simultaneous application of two or more different BEs, such as an adenine BE (which converts A·T base pairs to G·C) and a cytosine BE (which converts C·G base pairs to T·A), is not feasible because guide RNA crosstalk results in non-orthogonal editing, with all BEs modifying all target loci. Here we engineer both adenine BEs and cytosine BEs that can be orthogonally multiplexed by using RNA aptamer–coat protein systems to recruit the DNA-modifying enzymes directly to the guide RNAs. We generate four multiplexed orthogonal BE systems that enable rates of precise co-occurring edits of up to 7.1% in the same DNA strand without enrichment or selection strategies. The addition of a fluorescent enrichment strategy increases co-occurring edit rates up to 24.8% in human cells. These systems are compatible with expanded protospacer adjacent motif and high-fidelity Cas9 variants, function well in multiple cell types, have equivalent or reduced off-target propensities compared with their parental systems and can model disease-relevant point mutation combinations.