2025-02-06 カリフォルニア大学サンディエゴ校(UCSD)
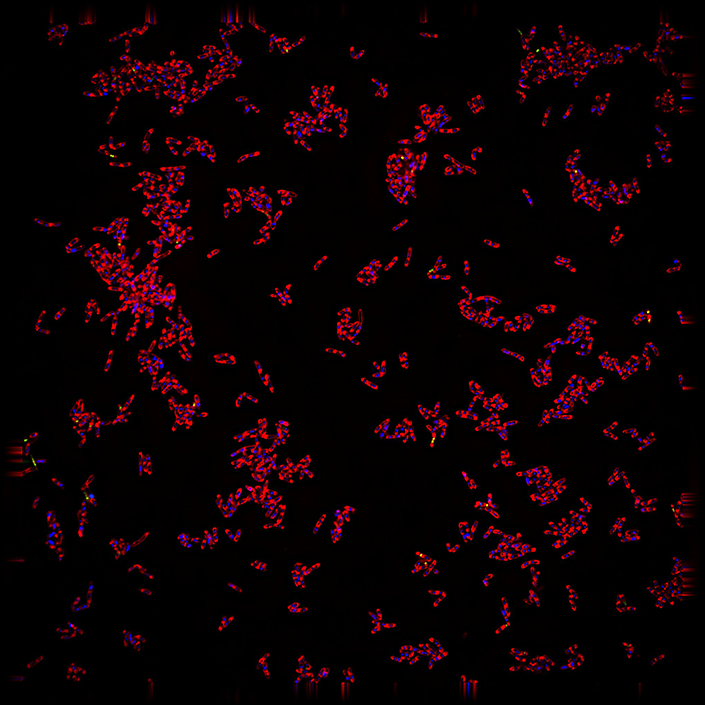
A fluorescence microscopy image reveals the tuberculosis-causing bacterium Mycobacterium tuberculosis after an antimicrobial treatment. Membranes are stained red, DNA blue and areas of membrane permeability appear green. These dramatic changes in bacterial cell structure form consistent patterns that can be used to identify how potential new treatments work — a critical step in developing effective therapies for the globally significant tuberculosis disease.
<関連情報>
- https://today.ucsd.edu/story/ai-accelerates-the-search-for-new-tuberculosis-drug-targets
- https://www.pnas.org/doi/10.1073/pnas.2419813122
結核菌の抗菌メカニズムを決定するためのディープラーニング駆動型細菌細胞学的プロファイリング Deep learning–driven bacterial cytological profiling to determine antimicrobial mechanisms in Mycobacterium tuberculosis
Diana Quach, Marc Sharp, Sara Ahmed, +5, and Joseph Sugie
Proceedings of the National Academy of Sciences Published:February 6, 2025
DOI:https://doi.org/10.1073/pnas.2419813122
Significance
Bacterial cytological profiling (BCP) is a well-established method to determine the mechanism of action (MOA) for antibiotics by examining the morphological changes that occur when bacteria are treated with a compound of interest. This study demonstrates the application of convolutional neural networks (CNN) to overcome technical challenges with a traditional approach to BCP, creating a robust platform to rapidly determine MOA for Mycobacterium tuberculosis. We demonstrate the capability of this platform by using it to confirm the MOA of several compounds that target M. tuberculosis. Our findings underscore the potential of CNN-based BCP to enhance the accuracy and efficiency of MOA determination, particularly for challenging pathogens like M. tuberculosis.
Abstract
Tuberculosis (TB), caused by Mycobacterium tuberculosis, remains a significant global health threat, affecting an estimated 10.6 million people in 2022. The emergence of multidrug resistant and extensively drug resistant strains necessitates the development of novel and effective drugs. Accelerating the determination of mechanisms of action (MOAs) for these drugs is crucial for advancing TB treatment. This study introduces MycoBCP, a unique adaptation of bacterial cytological profiling (BCP) tailored to M. tuberculosis, utilizing the application of convolutional neural networks (CNNs) within BCP to overcome challenges posed by traditional image analysis techniques. Using MycoBCP, we analyzed the morphological effects of various antimicrobial compounds on M. tuberculosis, capturing broad patterns rather than relying on precise cell segmentation. This approach circumvented issues such as cell clumping and uneven staining, which are prevalent in M. tuberculosis. In a blind test, MycoBCP accurately identified the MOA for 96% of the compounds, with a single misclassification of rifabutin, which was incorrectly categorized as affecting translation rather than transcription. The similar morphologies resulting from transcription and translation inhibition indicate a need for further refinement to distinguish them more effectively. Application of MycoBCP to a series of antitubercular agents successfully identified known MOAs and revealed unique effects, demonstrating its utility in early drug discovery and development. Our findings underscore the potential of CNN-based BCP to enhance the accuracy and efficiency of MOA determination, particularly for challenging pathogens like M. tuberculosis. MycoBCP represents a significant advancement in TB drug development, offering a robust and adaptable method for high-throughput screening of antimicrobial compounds.