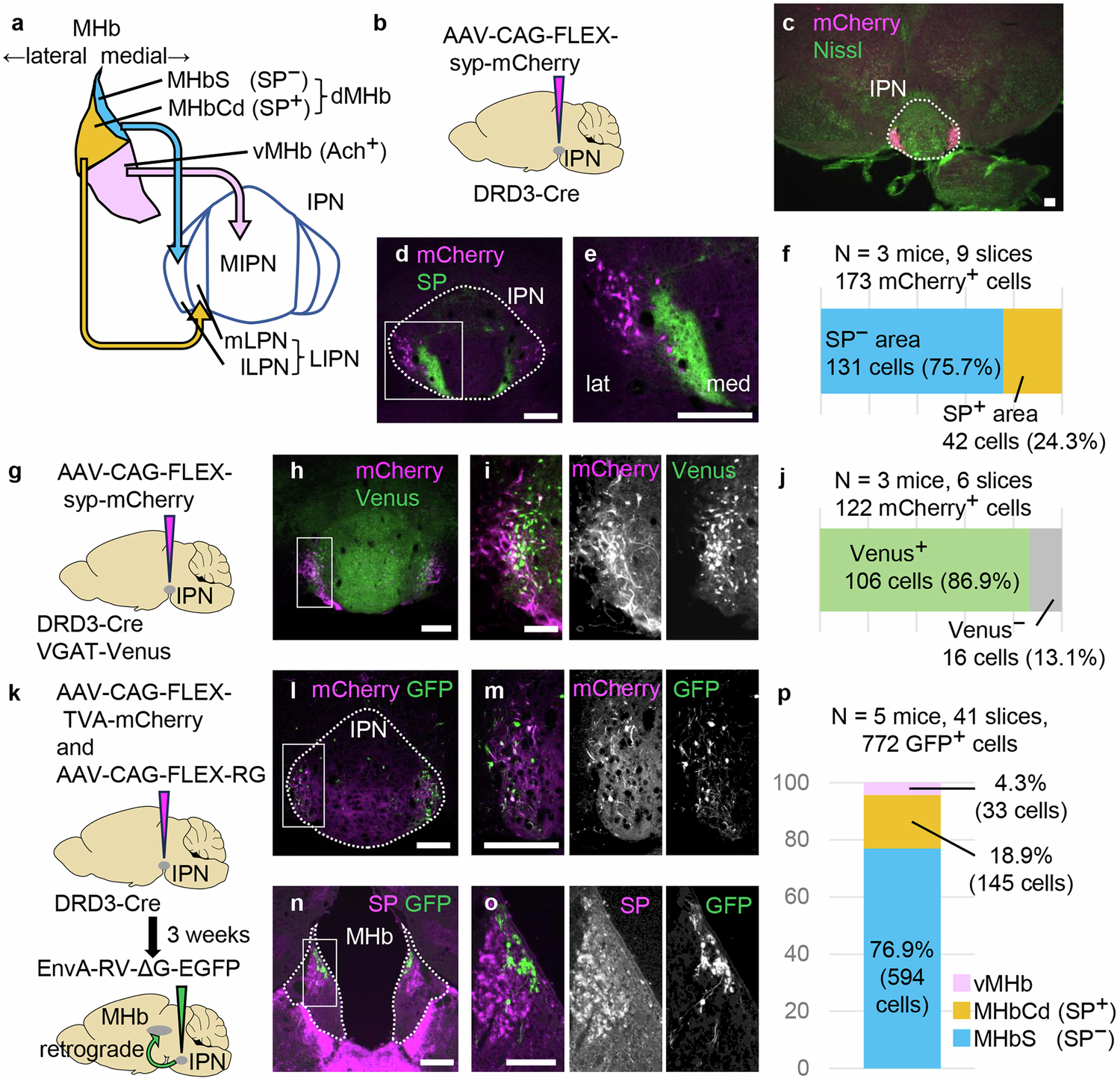
2025-03-31 国立遺伝学研究所
<関連情報>
- https://www.nig.ac.jp/nig/ja/2025/03/research-highlights_ja/pr20250329.html
- https://www.nig.ac.jp/nig/images/research_highlights/PR20250329.pdf
- https://www.science.org/doi/10.1126/sciadv.adu8400
複製依存的ヒストン標識が生きたヒト細胞におけるユークロマチン/ヘテロクロマチンの物理的性質を解明する Replication-dependent histone labeling dissects the physical properties of euchromatin/heterochromatin in living human cells
Katsuhiko Minami, Kako Nakazato, Satoru Ide, Kazunari Kaizu, […], and Kazuhiro Maeshima
Science Advances Published:28 Mar 2025
DOI:https://doi.org/10.1126/sciadv.adu8400

Abstract
A string of nucleosomes, where genomic DNA is wrapped around histones, is organized in the cell as chromatin, ranging from euchromatin to heterochromatin, with distinct genome functions. Understanding physical differences between euchromatin and heterochromatin is crucial, yet specific labeling methods in living cells remain limited. Here, we have developed replication-dependent histone (Repli-Histo) labeling to mark nucleosomes in euchromatin and heterochromatin based on DNA replication timing. Using this approach, we investigated local nucleosome motion in the four known chromatin classes, from euchromatin to heterochromatin, of living human and mouse cells. The more euchromatic (earlier-replicated) and more heterochromatic (later-replicated) regions exhibit greater and lesser nucleosome motions, respectively. Notably, the motion profile in each chromatin class persists throughout interphase. Genome chromatin is essentially replicated from regions with greater nucleosome motions, although the replication timing is perturbed. Our findings, combined with computational modeling, suggest that earlier-replicated regions have more accessibility, and local chromatin motion can be a major determinant of genome-wide replication timing.