2025-04-10 マックス・プランク研究所
<関連情報>
- https://www.mpg.de/24497100/a-new-tool-for-parsing-the-metabolic-dialogue-between-microorganisms
- https://www.nature.com/articles/s41467-025-58530-x
MetaFlowTrain: 代謝産物を介した生物間相互作用を研究するための高度に並列化されたモジュール式流体システム MetaFlowTrain: a highly parallelized and modular fluidic system for studying exometabolite-mediated inter-organismal interactions
Guillaume Chesneau,Johannes Herpell,Sarah Marie Wolf,Silvina Perin & Stéphane Hacquard
Nature Communications Published10 April 2025
DOIhttps://doi.org/10.1038/s41467-025-58530-x
Abstract
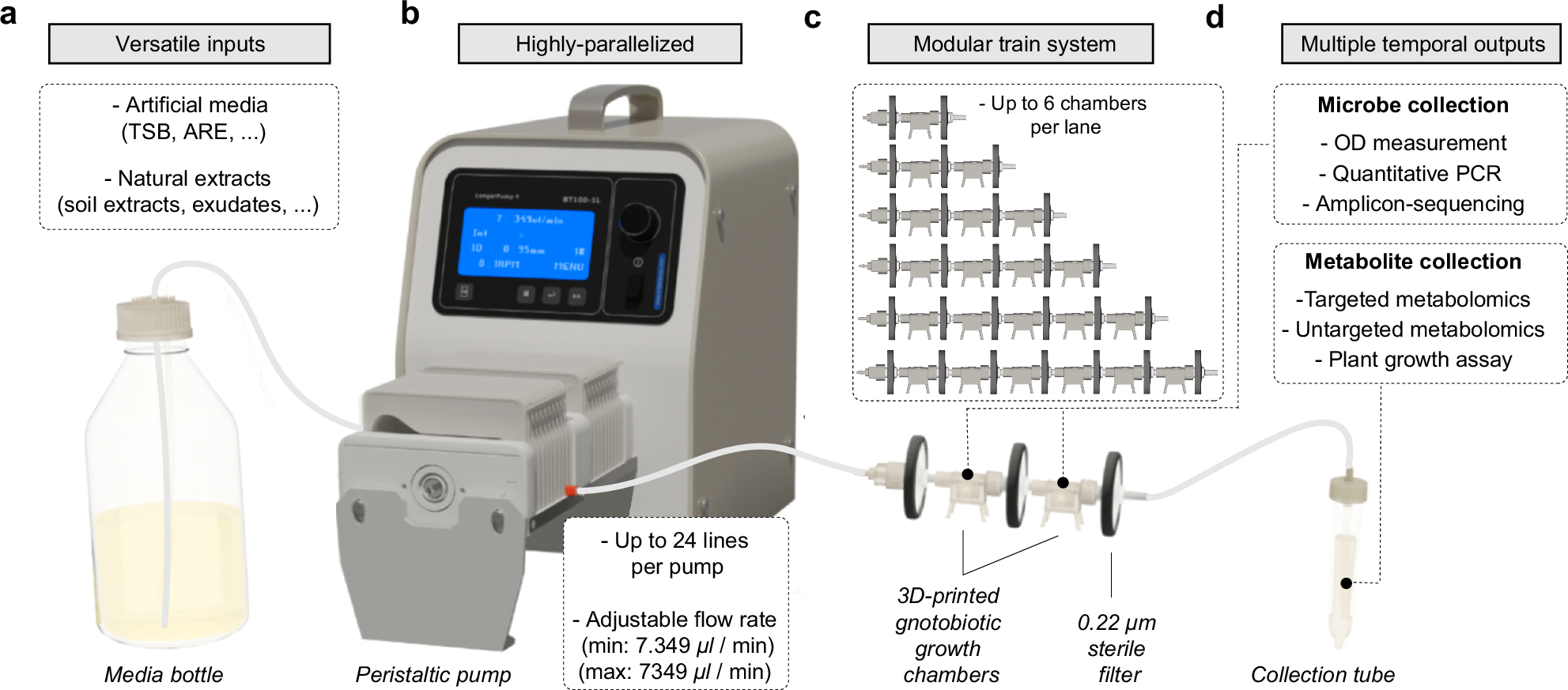
Metabolic fluxes between cells, organisms, or communities drive ecosystem assembly and functioning and explain higher-level biological organization. Exometabolite-mediated inter-organismal interactions, however, remain poorly described due to technical challenges in measuring these interactions. Here, we present MetaFlowTrain, an easy-to-assemble, cheap, semi-high-throughput, and modular fluidic system in which multiple media can be flushed at adjustable flow rates into gnotobiotic microchambers accommodating diverse micro-organisms, ranging from bacteria to small eukaryotes. These microchambers can be used alone or connected in series to create microchamber trains within which metabolites, but not organisms, directionally travel between microchambers to modulate organismal growth. Using MetaFlowTrain, we uncover soil conditioning effects on synthetic community structure and plant growth, and reveal microbial antagonism mediated by exometabolite production. Our study highlights MetaFlowTrain as a versatile system for investigating plant-microbe-microbe metabolic interactions. We also discuss the system´s potential to discover metabolites that function as signaling molecules, drugs, or antimicrobials across various systems.