2025-04-16 マックス・プランク研究所
<関連情報>
- https://www.mpg.de/24563841/0416-bioc-alpha-1-antitrypsin-deficiency-what-protects-the-one-and-not-the-other-153945-x
- https://www.nature.com/articles/s41586-025-08885-4
ディープ・ビジュアル・プロテオミクスが遺伝性肝疾患におけるタンパク質毒性をマッピング Deep Visual Proteomics maps proteotoxicity in a genetic liver disease
Florian A. Rosenberger,Sophia C. Mädler,Katrine Holtz Thorhauge,Sophia Steigerwald,Malin Fromme,Mikhail Lebedev,Caroline A. M. Weiss,Marc Oeller,Maria Wahle,Andreas Metousis,Maximilian Zwiebel,Niklas A. Schmacke,Sönke Detlefsen,Peter Boor,Ondřej Fabián,Soňa Fraňková,Aleksander Krag,Pavel Strnad & Matthias Mann
Nature Published:16 April 2025
DOI:https://doi.org/10.1038/s41586-025-08885-4
Abstract
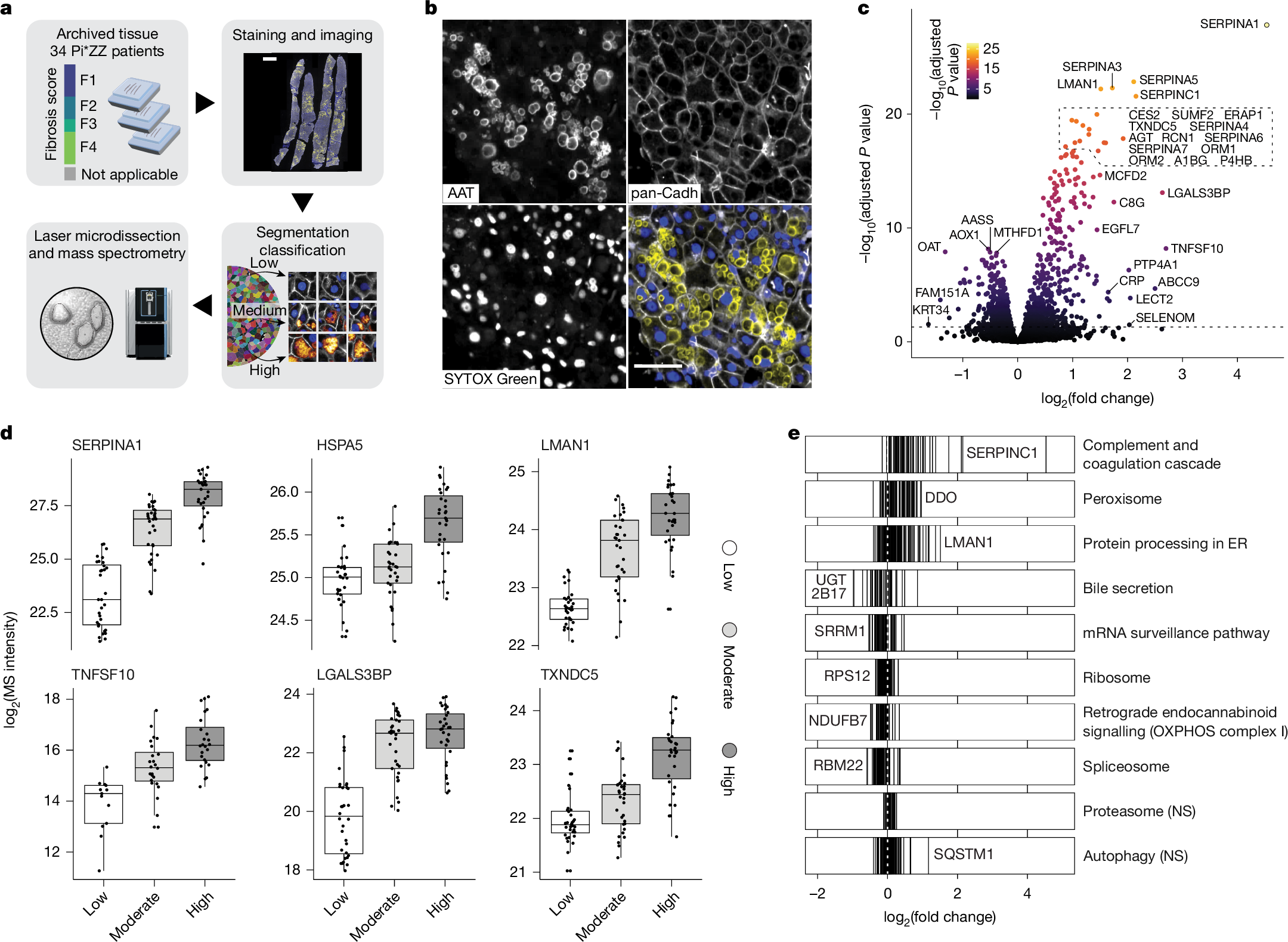
Protein misfolding diseases, including α1-antitrypsin deficiency (AATD), pose substantial health challenges, with their cellular progression still poorly understood1,2,3. We use spatial proteomics by mass spectrometry and machine learning to map AATD in human liver tissue. Combining Deep Visual Proteomics (DVP) with single-cell analysis4,5, we probe intact patient biopsies to resolve molecular events during hepatocyte stress in pseudotime across fibrosis stages. We achieve proteome depth of up to 4,300 proteins from one-third of a single cell in formalin-fixed, paraffin-embedded tissue. This dataset reveals a potentially clinically actionable peroxisomal upregulation that precedes the canonical unfolded protein response. Our single-cell proteomics data show α1-antitrypsin accumulation is largely cell-intrinsic, with minimal stress propagation between hepatocytes. We integrated proteomic data with artificial intelligence-guided image-based phenotyping across several disease stages, revealing a late-stage hepatocyte phenotype characterized by globular protein aggregates and distinct proteomic signatures, notably including elevated TNFSF10 (also known as TRAIL) amounts. This phenotype may represent a critical disease progression stage. Our study offers new insights into AATD pathogenesis and introduces a powerful methodology for high-resolution, in situ proteomic analysis of complex tissues. This approach holds potential to unravel molecular mechanisms in various protein misfolding disorders, setting a new standard for understanding disease progression at the single-cell level in human tissue.

