2025-05-19 パシフィック・ノースウェスト国立研究所(PNNL)
<関連情報>
- https://www.pnnl.gov/publications/gene-regulatory-networks-respond-day-and-night-cues-affect-phenotype-cyanbacteria
- https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2025.1569559/full
遺伝子ネットワーク中心性解析により、Synechococcus elongatus PCC 7942における昼夜の代謝転換を調整する重要な制御因子が同定された
Gene network centrality analysis identifies key regulators coordinating day-night metabolic transitions in Synechococcus elongatus PCC 7942 despite limited accuracy in predicting direct regulator-gene interactions
Zachary Johnson,David Anderson,Margaret S. Cheung,Pavlo Bohutskyi
Frontiers in Microbiology Published:26 March 2025
DOI:https://doi.org/10.3389/fmicb.2025.1569559
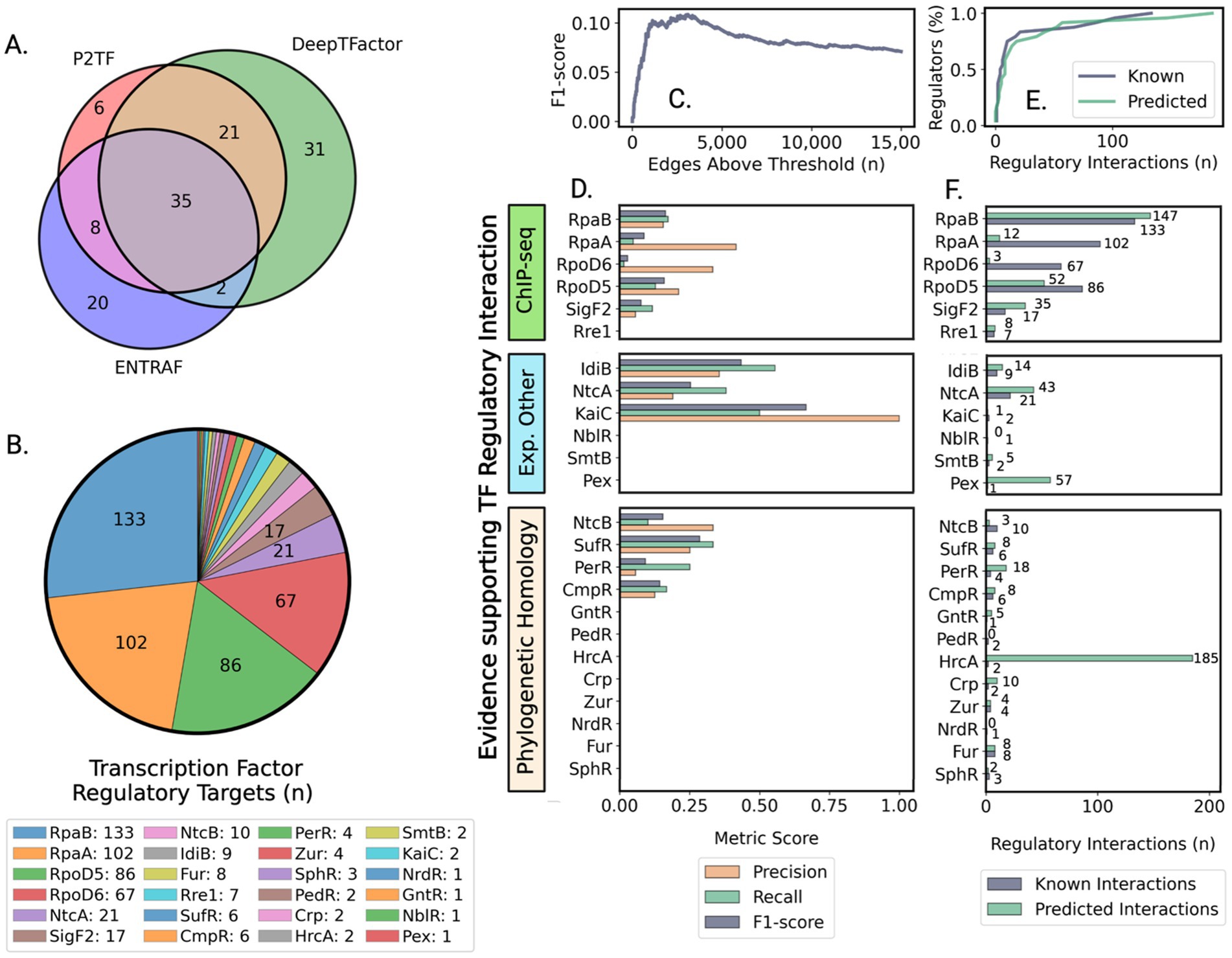
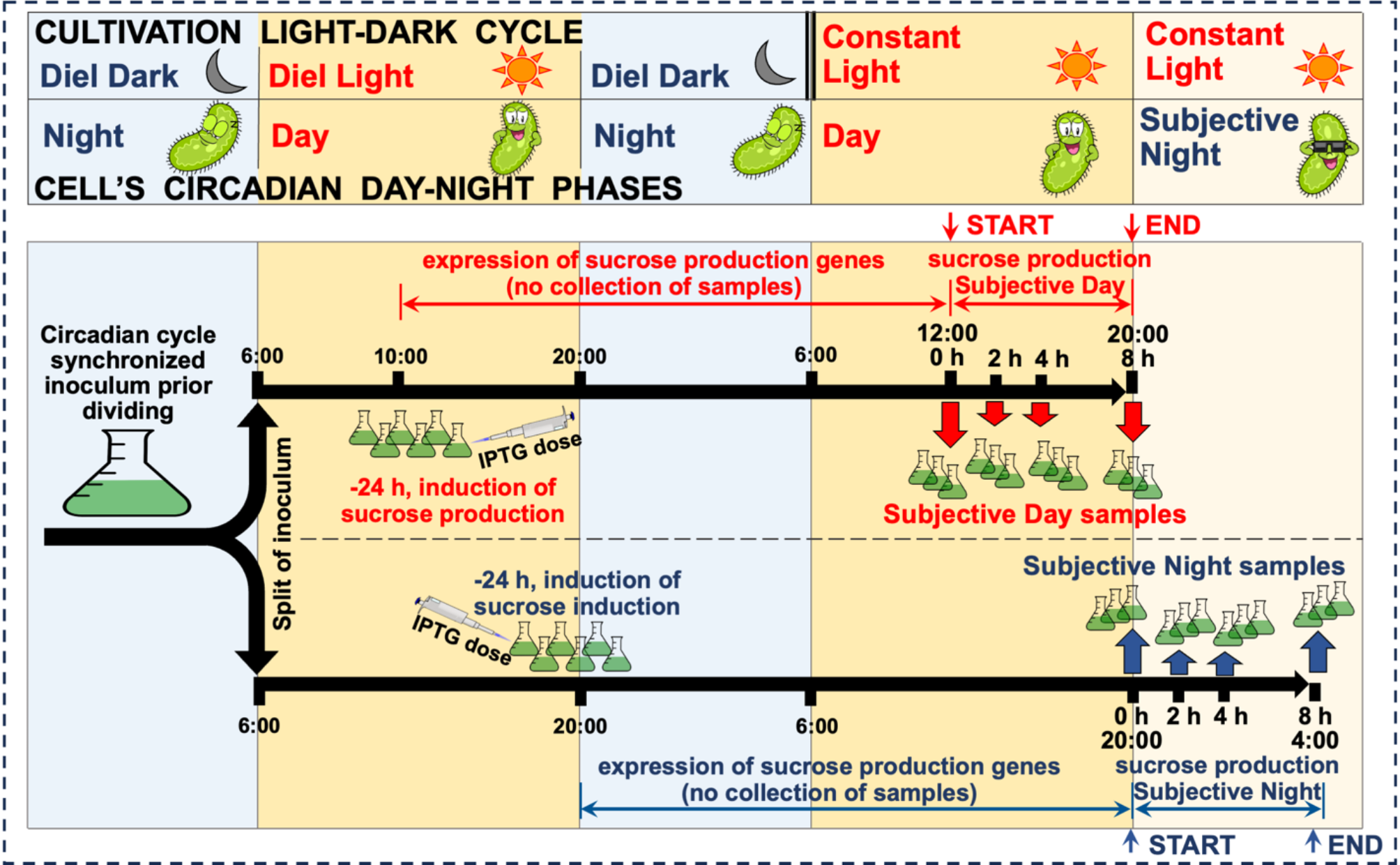
Synechococcus elongatus PCC 7942 is a model organism for studying circadian regulation and bioproduction, where precise temporal control of metabolism significantly impacts photosynthetic efficiency and CO2-to-bioproduct conversion. Despite extensive research on core clock components, our understanding of the broader regulatory network orchestrating genome-wide metabolic transitions remains incomplete. We address this gap by applying machine learning tools and network analysis to investigate the transcriptional architecture governing circadian-controlled gene expression. While our approach showed moderate accuracy in predicting individual transcription factor-gene interactions – a common challenge with real expression data – network-level topological analysis successfully revealed the organizational principles of circadian regulation. Our analysis identified distinct regulatory modules coordinating day-night metabolic transitions, with photosynthesis and carbon/nitrogen metabolism controlled by day-phase regulators, while nighttime modules orchestrate glycogen mobilization and redox metabolism. Through network centrality analysis, we identified potentially significant but previously understudied transcriptional regulators: HimA as a putative DNA architecture regulator, and TetR and SrrB as potential coordinators of nighttime metabolism, working alongside established global regulators RpaA and RpaB. This work demonstrates how network-level analysis can extract biologically meaningful insights despite limitations in predicting direct regulatory interactions. The regulatory principles uncovered here advance our understanding of how cyanobacteria coordinate complex metabolic transitions and may inform metabolic engineering strategies for enhanced photosynthetic bioproduction from CO2.