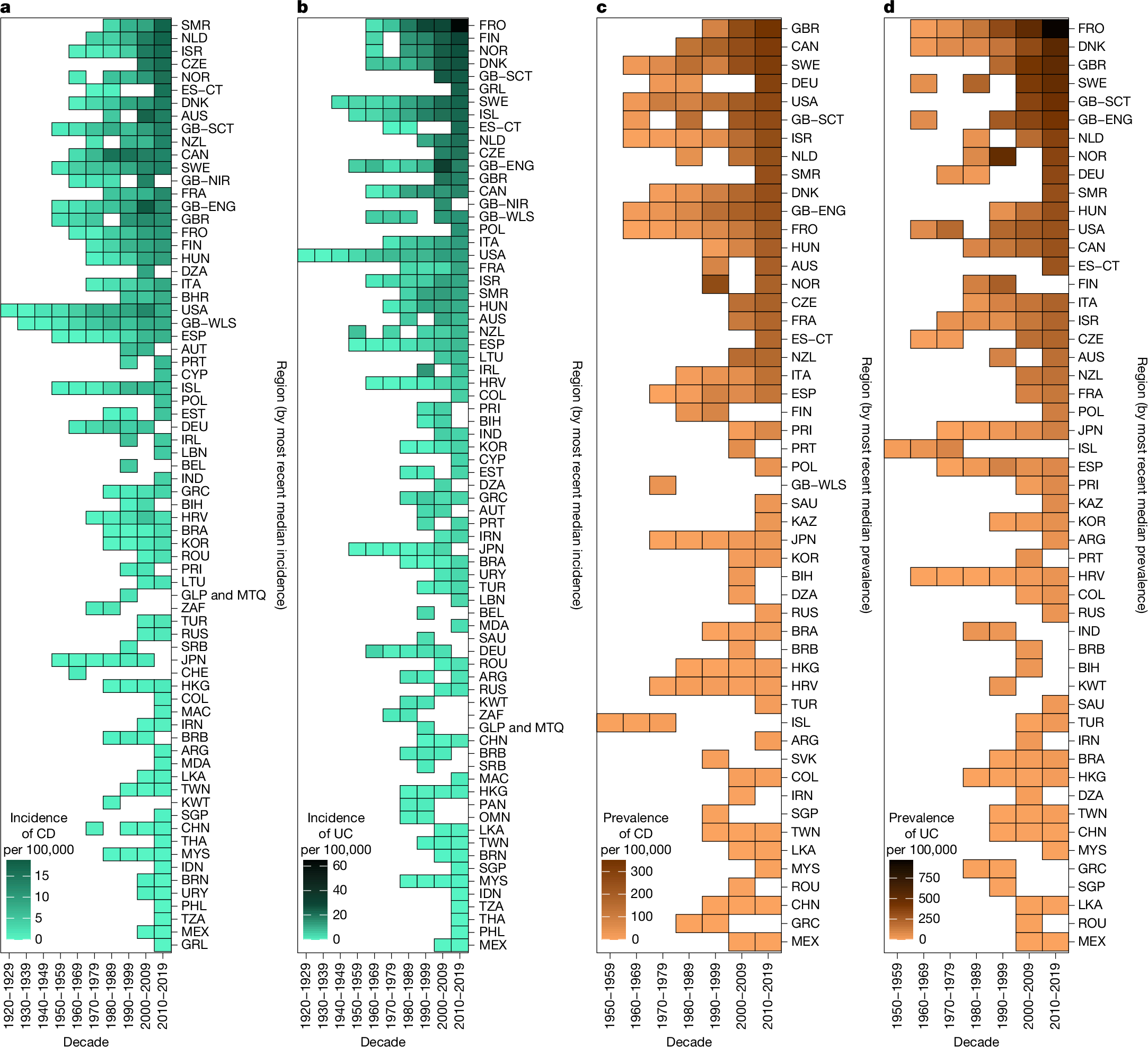
2025-08-01 タフツ大学
ChatGPT:

DNA repair enzymes, such as the ligase shown here that joins broken DNA strands, scan replicating DNA in the nucleus. Particularly difficult cases are now known to be transported to the inner edge of the nucleus, to be fixed by a different set of DNA repair enzymes. Image: Tom Ellenberger, National Institute of General Medical Science
<関連情報>
- https://now.tufts.edu/2025/08/01/cells-have-second-dna-repair-toolbox-difficult-cases
- https://www.sciencedirect.com/science/article/pii/S221112472500854X?via%3Dihub
DNA複製チェックポイントはキネトコアを標的とし、DNA構造による複製損傷を核周辺部に再配置する The DNA replication checkpoint targets the kinetochore to reposition DNA structure-induced replication damage to the nuclear periphery
Tyler M. Maclay, Jenna M. Whalen, Matthew J. Johnson, Catherine H. Freudenreich
Cell Reports Available online: 30 July 2025
DOI:https://doi.org/10.1016/j.celrep.2025.116083
Highlights
- The DNA replication checkpoint triggers repositioning of CAG/CTG tracts to the NPC
- Mrc1 phosphorylation requirement implicates fork uncoupling as the checkpoint signal
- Cep3 phosphorylation allows centromere release and is critical for NPC association
- Damage-inducible microtubules (DIMs) are required for repeat relocation to the NPC
Summary
Hairpin-forming CAG/CTG repeats pose significant challenges to DNA replication. In S. cerevisiae, long CAG/CTG repeat tracts reposition from the interior of the nucleus to the nuclear pore complex (NPC) to maintain their integrity. We show that relocation of a (CAG/CTG)130 tract to the NPC is dependent on phosphorylation of Mrc1 (hClaspin) of the fork protection complex and activation of the Mrc1/Rad53 replication checkpoint, implicating an uncoupled fork as the initial damage signal. Dun1-mediated phosphorylation of the kinetochore protein Cep3 is required for repositioning, a constraint that can be overcome by centromere inactivation, connecting detachment of the kinetochore from microtubule ends to NPC association. Activation of this pathway leads to the formation of DNA damage-induced microtubules, which associate with the repeat and are necessary for locus repositioning. These data implicate the replication checkpoint in facilitating the movement of DNA structure-associated damage to the nuclear periphery by centromere release and microtubule-directed motion.