2025-08-11 医薬基盤・健康・栄養研究所
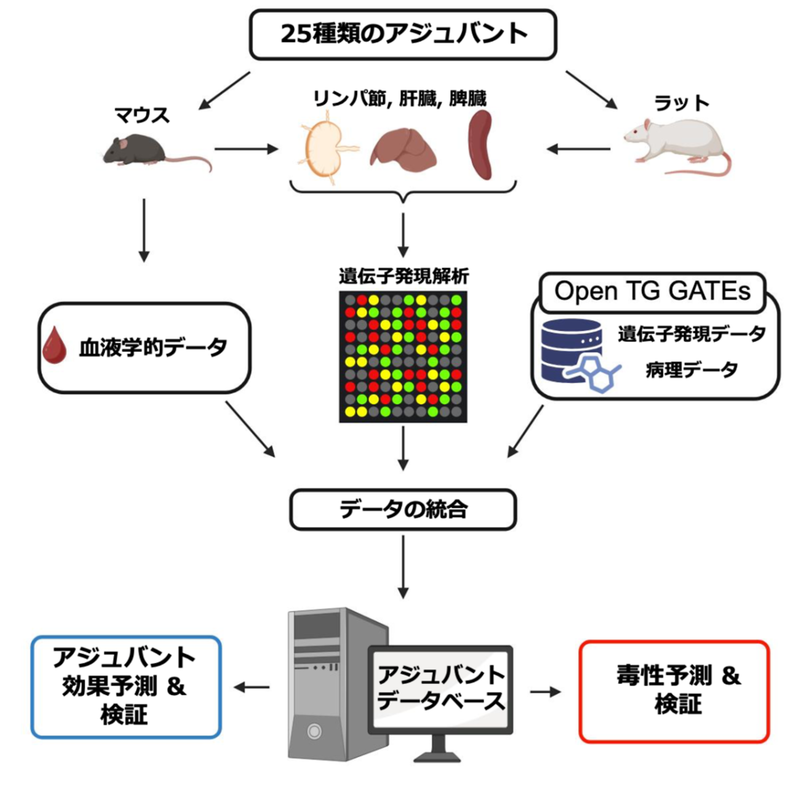
図1:アジュバントデータベース(ADB)の構築と解析
<関連情報>
- https://www.nibn.go.jp/pr/press/20250811.html
- https://www.nibn.go.jp/pr/press/documents/20250811_2.pdf
- https://www.cell.com/cell-chemical-biology/fulltext/S2451-9456(25)00228-4
ワクチンと免疫療法の非臨床評価用の補助データベース An adjuvant database for preclinical evaluation of vaccines and immunotherapeutics
Yayoi Natsume-Kitatani ∙ Kouji Kobiyama ∙ Yoshinobu Igarashi ∙ … ∙ Hiroshi Yamada ∙ Kenji Mizuguchi ∙ Ken J. Ishii
Cell Chemical Biology Published:August 11, 2025
DOI:https://doi.org/10.1016/j.chembiol.2025.07.005
Significance
Adjuvants are vital components of vaccine formulations that enhance immune responses and hold therapeutic potential beyond prophylaxis, including applications in cancer immunotherapy and infectious disease treatment. However, systematic evaluation of their efficacy and safety remains challenge due to the lack of standardized, high-throughput screening platforms. Current methods are often limited by biological variability and inconsistent experimental conditions that hinder reproducibility and scalability in adjuvant discovery.
To address this bottleneck, we developed the Adjuvant Database (ADB), a prototype multi-species transcriptome resource constructed using harmonized protocols similar to those employed in Open TG-GATEs (OTG), a widely used toxicogenomics database. ADB contains comprehensive gene expression profiles linked to physiological measurements for 25 core adjuvants across multiple organs, time points, doses, and species. The use of standardized protocols enables integrative analyses with OTG, allowing for simultaneous prediction of both immunostimulatory efficacy (adjuvanticity) and potential toxicity (e.g., hepatotoxicity).
Our machine learning analyses demonstrated that transcriptomic signatures are robust enough to identify individual adjuvants regardless of organ or species. Additionally, we successfully built predictive models for adjuvanticity and hepatotoxicity, revealing novel immunostimulatory properties of colchicine and the hepatotoxicity of FK565 through a fully data-driven approach.
The integration of ADB with existing toxicogenomic resources like OTG marks a significant step toward rational, transcriptomic-based adjuvant screening. This framework not only accelerates the identification of promising candidates but also provides a scalable, standardized platform for safety evaluation. Ultimately, this approach will facilitate the development of safer and more effective immunotherapeutics by leveraging systems-level transcriptomic data.
Highlights
- A transcriptomic database of 25 vaccine adjuvants was constructed
- Characteristics of adjuvants were reflected in gene expression profiles
- Integration with Open TG-GATEs enables prediction of toxicity and adjuvanticity
- The predictions by machine learning were validated by experiments
Summary
Adjuvants are immunostimulators used to enhance vaccine efficacy against infectious diseases. However, current methods for evaluating their efficacy and safety are limited, hindering large-scale screening. To address this, we developed a prototype Adjuvant Database (ADB) containing transcriptome data, generated using the same protocols as the widely used Open TG-GATEs (OTG) toxicogenomics database, covering 25 adjuvants across multiple species, organs, time points, and doses. This enabled cross-database integration of ADB and OTG. Transcriptomic patterns successfully distinguished each adjuvant regardless of organs or species. Using both databases, we built machine learning models to predict adjuvanticity and hepatotoxicity. Notably, we identified colchicine’s adjuvant activity and FK565’s liver toxicity through data-driven analysis. Overall, ADB combined with OTG offers a framework for transcriptomics-based, data-driven screening of adjuvant candidates.

