2025-09-09 ペンシルベニア州立大学(Penn State)
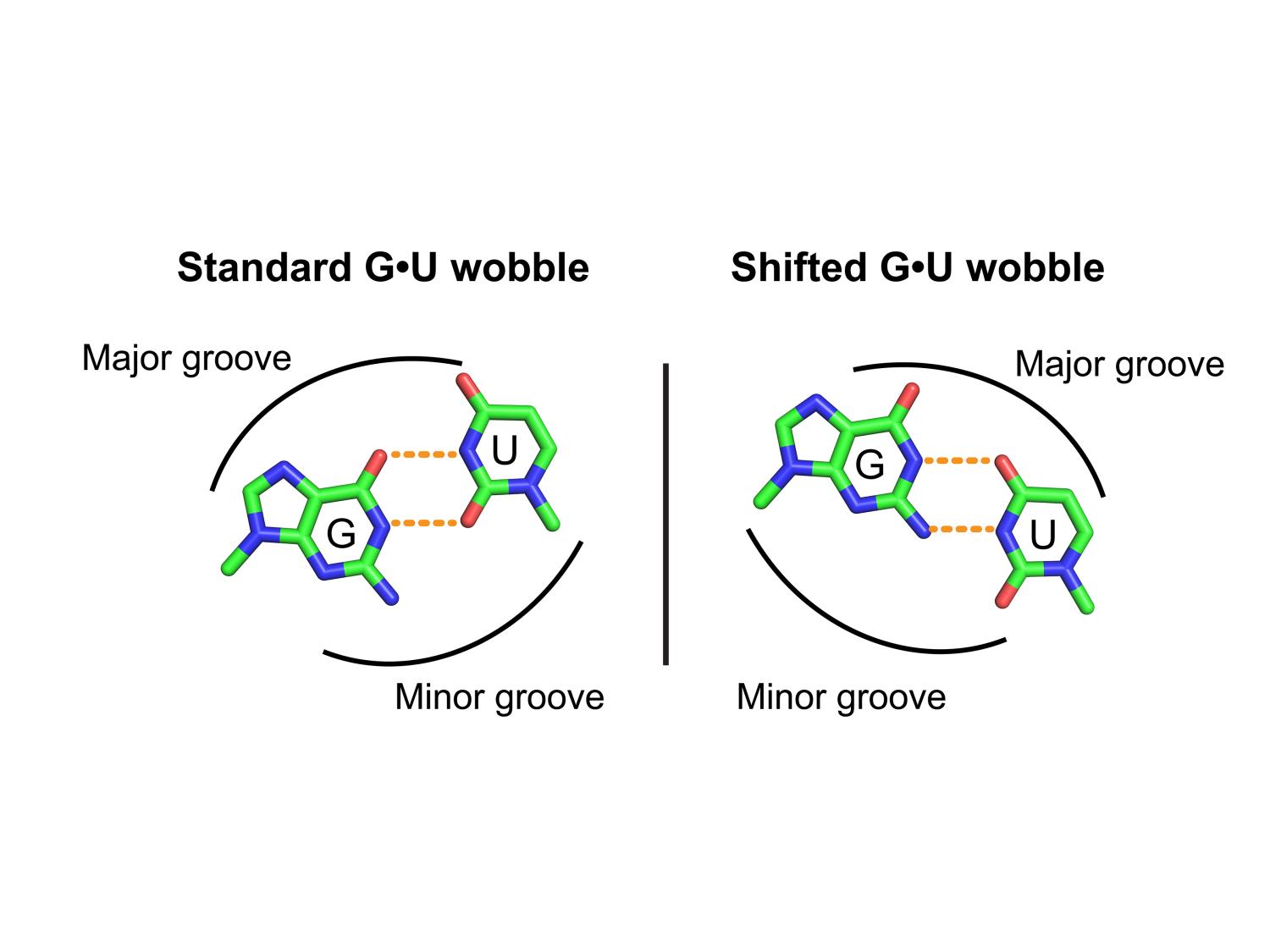 When the sidechain bases guanine (G) and uracil (U) are paired in the 3D structure of a molecule of ribonucleic acid (RNA) the unusual pairing creates an unusual molecular conformation called a “wobble,” pictured on the left. New research shows that non-covalent modifications to the bases can further alter the conformation creating a “shifted wobble,” shown on the right. This conformational diversity could help explain RNAs function versatility, according to the researchers. Credit: Md Sharear Saon/Bevilacqua Lab / Penn State. Creative Commons
When the sidechain bases guanine (G) and uracil (U) are paired in the 3D structure of a molecule of ribonucleic acid (RNA) the unusual pairing creates an unusual molecular conformation called a “wobble,” pictured on the left. New research shows that non-covalent modifications to the bases can further alter the conformation creating a “shifted wobble,” shown on the right. This conformational diversity could help explain RNAs function versatility, according to the researchers. Credit: Md Sharear Saon/Bevilacqua Lab / Penn State. Creative Commons
<関連情報>
- https://www.psu.edu/news/eberly-college-science/story/unusual-molecular-conformation-could-help-explain-rnas-versatility
- https://academic.oup.com/nar/article/53/14/gkaf575/8210589?login=false
RNAの代替プロトン化に起因するシフトしたG・Uウォブル対の同定と特性評価 Identification and characterization of shifted G•U wobble pairs resulting from alternative protonation of RNA
Md Sharear Saon, Catherine A Douds, Andrew J Veenis, Ashley N Pearson, Neela H Yennawar, Philip C Bevilacqua
Nucleic Acids Research Published:22 July 2025
DOI:https://doi.org/10.1093/nar/gkaf575
Abstract
RNA can serve as an enzyme, small molecule sensor, and vaccine, and it may have been a conduit for the origin of life. Despite these profound functions, RNA is thought to have limited molecular diversity. A pressing question is whether RNA can adopt novel molecular states that enhance its function. Covalent modifications of RNA have been demonstrated to augment biological function, but much less is known about non-covalent alterations such as novel protonated or tautomeric forms. Conventionally, a G•U wobble has the U located in the major groove. We used a cheminformatic approach to identify four structural families of shifted G•U wobbles in which the G instead resides in the major groove, which requires alternative tautomeric states of either base, or an anionic state of the U. We provide experimental support for these shifted G•U wobbles via the unconventional in vivo reactivity of the U with dimethylsulfate (DMS). These shifted wobbles may play functional roles and could serve as drug targets, as they are common in Bacteria and chloroplasts, but underrepresented in Eukaryotes and Archaea. Our cheminformatics approach can be applied to identify alternative protonation states in other RNA motifs, as well as in DNA and proteins.

