2024-03-26 カリフォルニア大学サンディエゴ校(UCSD)
<関連情報>
- https://today.ucsd.edu/story/new-genetic-analysis-tool-tracks-risks-tied-to-crispr-edits
- https://link.springer.com/article/10.1038/s41467-024-46479-2
DNA二本鎖切断修復の発生過程をシングルアレルの分解能を持つ変異分類装置で読み解く Developmental progression of DNA double-strand break repair deciphered by a single-allele resolution mutation classifier
Zhiqian Li,Lang You,Anita Hermann & Ethan Bier
Nature Communications Published:23 March 2024
DOI:https://doi.org/10.1038/s41467-024-46479-2
Abstract
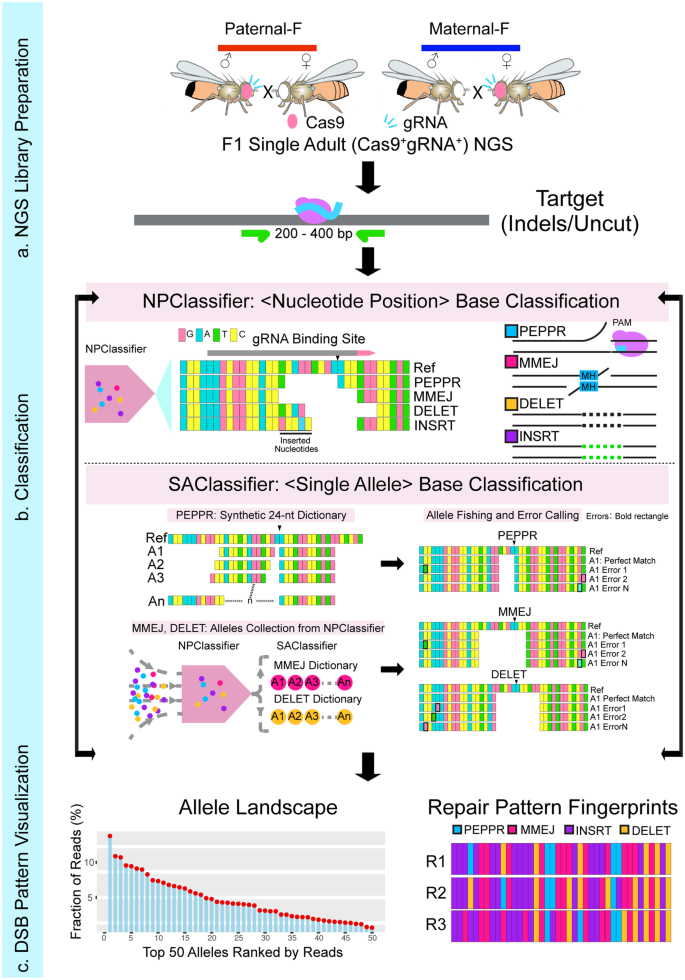
DNA double-strand breaks (DSBs) are repaired by a hierarchically regulated network of pathways. Factors influencing the choice of particular repair pathways, however remain poorly characterized. Here we develop an Integrated Classification Pipeline (ICP) to decompose and categorize CRISPR/Cas9 generated mutations on genomic target sites in complex multicellular insects. The ICP outputs graphic rank ordered classifications of mutant alleles to visualize discriminating DSB repair fingerprints generated from different target sites and alternative inheritance patterns of CRISPR components. We uncover highly reproducible lineage-specific mutation fingerprints in individual organisms and a developmental progression wherein Microhomology-Mediated End-Joining (MMEJ) or Insertion events predominate during early rapid mitotic cell cycles, switching to distinct subsets of Non-Homologous End-Joining (NHEJ) alleles, and then to Homology-Directed Repair (HDR)-based gene conversion. These repair signatures enable marker-free tracking of specific mutations in dynamic populations, including NHEJ and HDR events within the same samples, for in-depth analysis of diverse gene editing events.