2025-12-19 ワシントン大学セントルイス校
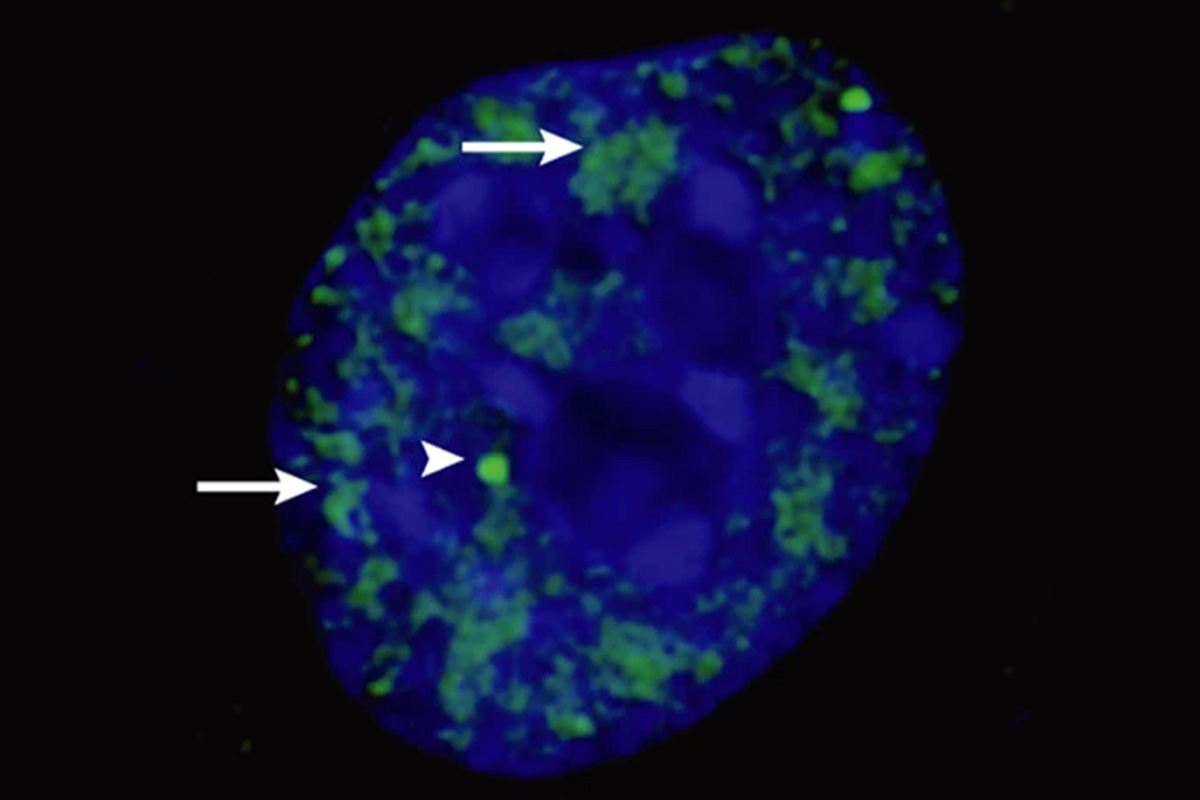
![Rohit V. Pappu, the Gene K. Beare Distinguished Professor of Biomedical Engineering, and Min Kyung Shinn, a former postdoctoral researcher in his lab, investigated some of the physical features of nuclear speckles that were inconsistent with the type of uniform droplets one expected to see if they formed via liquid-liquid phase separation. Their microscopy images revealed that the speckle-associated proteins were distinct in the types of structures they formed. [Credit: Nature Reviews Molecular Cell Biology 4, 605–612 (2003).]](https://engineering.washu.edu/news/images/rohit-image.jpg)
Credit: Nature Reviews Molecular Cell Biology 4, 605–612 (2003).
<関連情報>
- https://engineering.washu.edu/news/2025/Microphases-are-targets-for-therapeutic-intervention-for-neurodegenerative-disorders.html
- https://www.cell.com/cell/fulltext/S0092-8674(25)01361-3
核スペックルタンパク質は内因性およびMALAT1依存性のミクロ相を形成する Nuclear speckle proteins form intrinsic and MALAT1-dependent microphases
Min Kyung Shinn ∙ Dylan T. Tomares ∙ Vicky Liu ∙ … ∙ Matthew D. Lew ∙ Kannanganattu V. Prasanth ∙ Rohit V. Pappu
Cell Published:December 19, 2025
DOI:https://doi.org/10.1016/j.cell.2025.11.026
Highlights
- Proteins with RRMs and IDRs are defined by inter-domain attractions and repulsions
- Interplay of attractions and repulsions yields 25–40 nm protein-specific microphases
- MALAT1 binds preferentially to microphases by releasing positively charged proteins
- Microphases form clusters in cells and micron-scale core-shell emulsions in mixtures
Summary
Pre-mRNA processing components in nuclear speckles encompass one or more folded RNA recognition motifs (RRMs) and disordered regions with specific sequence grammars. Such proteins include serine/arginine-rich splicing factors (SRSFs) and transactive response DNA binding protein (TDP)-43. The SRSFs and TDP-43 are unique archetypes of block copolymers encoding specific patterns of inter-domain homotypic and heterotypic attractions and repulsions. The interplay of these interactions drives microphase separation and the formation of ordered, size-limited assemblies. Microphases of SRSFs and TDP-43 are 23–45 nm in diameter, each comprising tens of molecules. Sub-micron-scale assemblies of SRSFs in cells are consistent with being clusters of microphases. The speckle-associated regulatory long non-coding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) binds specifically and preferentially to SRSF1 microphases, while destabilizing TDP-43 microphases. In protein mixtures, the interactions between microphases drive the formation of micron-scale double-emulsion structures with core-shell organization. Our findings show how interactions involving copolymers featuring folded domains and disordered regions drive the formation of microphases.