2026-01-21 医薬基盤・健康・栄養研究所
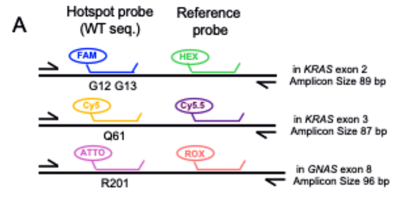
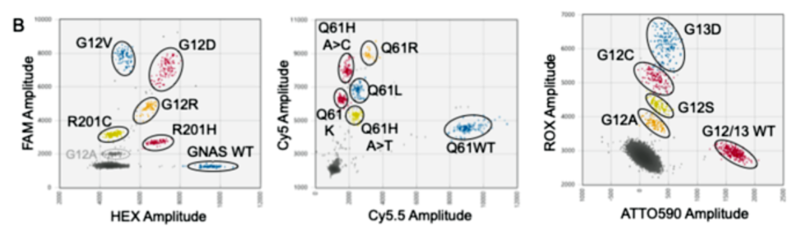
図1.PlexScreen-dPCR解析によるKRAS、GNASのスクリーニング法(A:プローブ構成の模式図、B:各ドライバー変異および野生型のクラスターシフトパターン)
<関連情報>
- https://www.nibn.go.jp/pr/press/20260121.html
- https://www.nibn.go.jp/pr/press/documents/20260121.pdf
- https://academic.oup.com/clinchem/advance-article/doi/10.1093/clinchem/hvaf181/8418283
膵がん発生に関連する複数のドライバー変異のスクリーニングおよび変異同定のための2つの6色マルチプレックスデジタルPCR Six-Color Multiplex Digital PCR Assays for Comprehensive Screening and Identification of Multiple Driver Mutations Associated with Pancreatic Carcinogenesis
Chiho Maeda,Yusuke Ono,Kenji Takahashi,Miyuki Mori,Mayumi Suzuki,Nobue Tamamura,Yanpeng Sun,Taito Itoh,Hiroki Tanaka,Hidemasa Kawabata
Clinical Chemistry Published:09 January 2026
DOI:https://doi.org/10.1093/clinchem/hvaf181
Abstract
Background
Digital polymerase chain reaction (dPCR) is widely recognized for its high sensitivity in detecting low-frequency variants; however, conventional 2-color systems have limited multiplex capacity. Expanding this capability is essential for simultaneous detection of multiple driver mutations in cancer-related genes. KRAS and GNAS are key driver genes in the early development of pancreatic cancer and its precursor lesions, and mutations in these genes are often present at low abundance in clinical samples.
Methods
Two 6-color dPCR assays were developed using a droplet-based platform. PlexScreen-dPCR is a multicolored drop-off assay designed to screen for mutations in KRAS codons 12/13 and 61 and GNAS codon 201, without specifying individual variants. PlexID-dPCR employs variant-specific probes to distinguish among 14 predefined KRAS and GNAS mutations in a single reaction. The assays were validated using synthetic DNA, cell lines, 23 tissue samples, and 12 duodenal fluid samples. Customized primer/probe sets with 6 fluorophores were employed in a 6-color droplet dPCR system, and the limits of detection (LOD) were determined.
Results
PlexScreen-dPCR, applied in contrived samples, demonstrated LODs as low as 0.03% to 0.06%, enabling high-sensitivity detection of low-abundance mutations. PlexID-dPCR accurately identified all 14 variants in a single well. Both assays showed complete concordance with conventional methods, exhibiting a strong correlation for variant allele frequency quantification.
Conclusions
These 6-color dPCR assays offer scalable solutions for improved throughput detection of KRAS and GNAS mutations. Their compatibility with commercially available platforms and streamlined workflow support their integration into clinical practice. Further optimization can enhance cluster interpretation in high-plex settings and facilitate expansion toward broader genomic targets.
