2026-02-06 清華大学
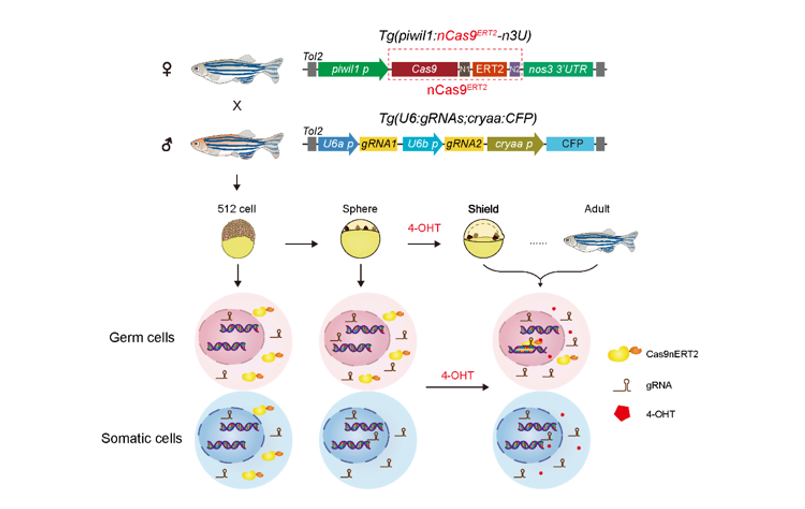
The schematic diagram of the zebrafish spatiotemporally inducible genome editing system and transgenic zebrafish lines.
<関連情報>
- https://www.tsinghua.edu.cn/en/info/1245/14707.htm
- https://rupress.org/jcb/article/225/4/e202412216/281346/Stage-and-tissue-specific-gene-editing-using-4-OHT
4-OHT誘導性Cas9を用いた全生物における段階および組織特異的な遺伝子編集
Stage- and tissue-specific gene editing using 4-OHT–inducible Cas9 in whole organism
Yaqi Li,Weiying Zhang,Zihang Wei,Han Li,Xin Liu,Tao Zheng,Tursunjan Aziz,Cencan Xing,Anming Meng,Xiaotong Wu
Journal of Cell Biology Published:January 02 2026
DOI:https://doi.org/10.1083/jcb.202412216
Vertebrate genes function in specific tissues and stages, so their functional studies require conditional knockout or editing. In zebrafish, spatiotemporally inducible genome editing, particularly during early embryogenesis, remains challenging. Here, we establish inducible Cas9-based editing in defined cell types and stages. The nCas9ERT2 fusion protein, consisting of Cas9 and an estrogen receptor flanked by two nuclear localization signals, is usually located in the cytoplasm and efficiently translocated into nuclei upon 4-hydroxytamoxifen (4-OHT) treatment in cultured cells or embryos. As a proof of concept, we demonstrate that genes in primordial germ cells in embryos and germ cells in adult ovaries from a transgenic line with stable expression of nCas9ERT2 and gRNAs can be mutated by 4-OHT induction. The system also works in early mouse embryos. Thus, this inducible nCas9ERT2 approach enables temporospatial gene editing at the organismal level, expanding the tissue- and stage-specific gene-editing toolkit.

.jpg)