2022-08-17 スイス連邦工科大学ローザンヌ校(EPFL)
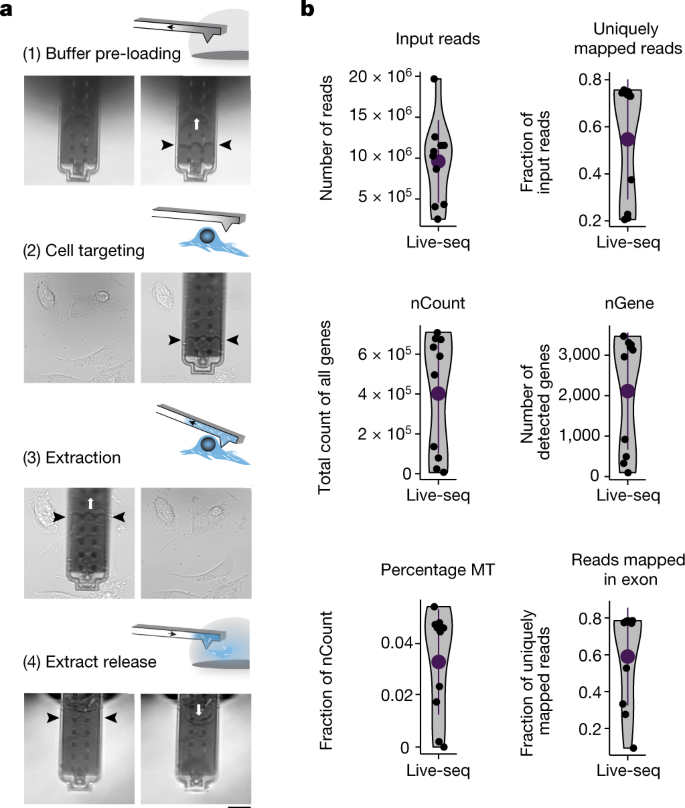
Live-seqの鍵となるのは、「流体力顕微鏡」またはFluidFMと呼ばれる顕微鏡技術で、人間の髪の毛よりも細い微小なチャンネルを使って、数年前にチューリッヒ工科大学が開発した顕微鏡下でサンプル中の微量(フェムトリットル)流体を操作するものです。そのため、FluidFMでは、個々の細胞に物質を挿入したり、細胞を殺さずにmRNAを含む細胞質を取り出したりすることができる。
Live-seqによって、ある時点の細胞の微量の細胞質サンプルからmRNA(トランスクリプトーム)を読み出し、その後の分子的・表現的挙動と結びつけることができるようになった。
<関連情報>
- https://actu.epfl.ch/news/live-seq-sequencing-a-cell-without-killing-it/
- https://www.nature.com/articles/s41586-022-05046-9
ライブセクにより単一細胞のトランスクリプトーム経時的記録を実現 Live-seq enables temporal transcriptomic recording of single cells
Wanze Chen,Orane Guillaume-Gentil,Pernille Yde Rainer,Christoph G. Gäbelein,Wouter Saelens,Vincent Gardeux,Amanda Klaeger,Riccardo Dainese,Magda Zachara,Tomaso Zambelli,Julia A. Vorholt & Bart Deplancke
Nature Published:17 August 2022
DOI:https://doi.org/10.1038/s41586-022-05046-9
Abstract
Single-cell transcriptomics (scRNA-seq) has greatly advanced our ability to characterize cellular heterogeneity1. However, scRNA-seq requires lysing cells, which impedes further molecular or functional analyses on the same cells. Here, we established Live-seq, a single-cell transcriptome profiling approach that preserves cell viability during RNA extraction using fluidic force microscopy2,3, thus allowing to couple a cell’s ground-state transcriptome to its downstream molecular or phenotypic behaviour. To benchmark Live-seq, we used cell growth, functional responses and whole-cell transcriptome read-outs to demonstrate that Live-seq can accurately stratify diverse cell types and states without inducing major cellular perturbations. As a proof of concept, we show that Live-seq can be used to directly map a cell’s trajectory by sequentially profiling the transcriptomes of individual macrophages before and after lipopolysaccharide (LPS) stimulation, and of adipose stromal cells pre- and post-differentiation. In addition, we demonstrate that Live-seq can function as a transcriptomic recorder by preregistering the transcriptomes of individual macrophages that were subsequently monitored by time-lapse imaging after LPS exposure. This enabled the unsupervised, genome-wide ranking of genes on the basis of their ability to affect macrophage LPS response heterogeneity, revealing basal Nfkbia expression level and cell cycle state as important phenotypic determinants, which we experimentally validated. Thus, Live-seq can address a broad range of biological questions by transforming scRNA-seq from an end-point to a temporal analysis approach.