2023-02-06 サセックス大学
◆癌の治療法は、主に、癌の部位と種類に基づいて処方されます。腫瘍の遺伝的な違いによって、標準的ながん治療が効かなくなることがあります。治療の指針として個別化されたアプローチを用いることで、がん患者の平均余命や生活の質を向上させ、不必要な副作用を減らすことができるかもしれません。
<関連情報>
- https://www.sussex.ac.uk/research/full-news-list?id=59950
- https://academic.oup.com/bioinformaticsadvances/article/2/1/vbac084/6820950
生物学的ネットワークトポロジーの特徴から、がん細胞株における遺伝子依存性を予測する。 Biological network topology features predict gene dependencies in cancer cell-lines
Graeme Benstead-Hume, Sarah K Wooller, Joanna Renaut, Samantha Dias, Lisa Woodbine, Antony M Carr, Frances M G Pearl
Bioinformatics Advances Published:10 November 2022
DOI:https://doi.org/10.1093/bioadv/vbac084
Abstract
Motivation
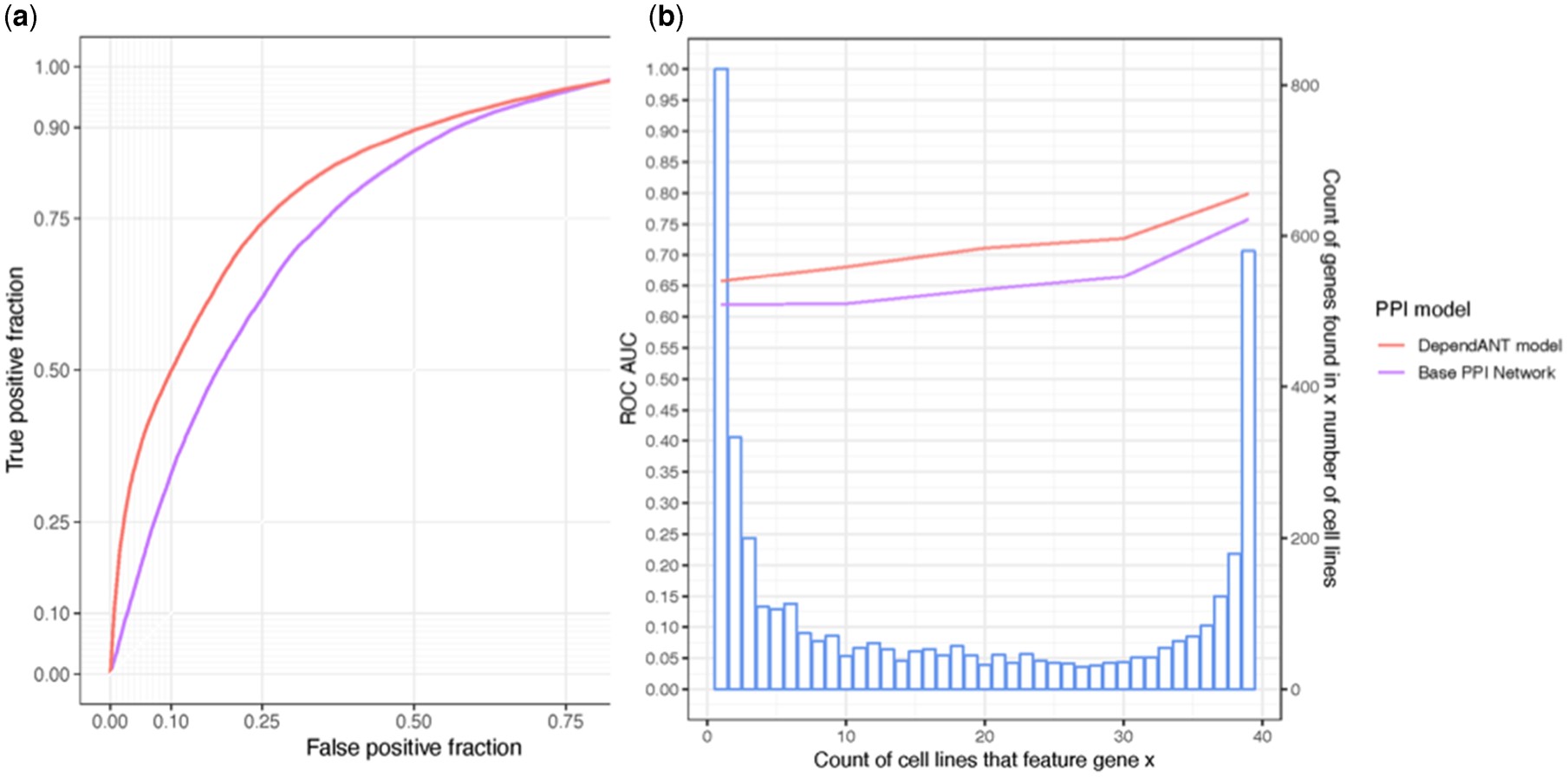
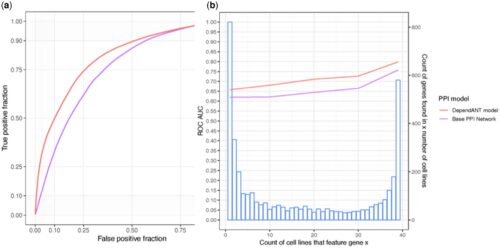
Protein–protein interaction (PPI) networks have been shown to successfully predict essential proteins. However, such networks are derived generically from experiments on many thousands of different cells. Consequently, conventional PPI networks cannot capture the variation of genetic dependencies that exists across different cell types, let alone those that emerge as a result of the massive cell restructuring that occurs during carcinogenesis. Predicting cell-specific dependencies is of considerable therapeutic benefit, facilitating the use of drugs to inhibit those proteins on which the cancer cells have become specifically dependent. In order to go beyond the limitations of the generic PPI, we have attempted to personalise PPI networks to reflect cell-specific patterns of gene expression and mutation. By using 12 topological features of the resulting PPIs, together with matched gene dependency data from DepMap, we trained random-forest classifiers (DependANT) to predict novel gene dependencies.
Results
We found that DependANT improves the power of the baseline generic PPI models in predicting common gene dependencies, by up to 10.8% and is more sensitive than the baseline generic model when predicting genes on which only a small number of cell types are dependent.
Availability and implementation
Software available at https://bitbucket.org/bioinformatics_lab_sussex/dependant2
Supplementary information
Supplementary data are available at Bioinformatics Advances online.