2023-03-15 カロリンスカ研究所(KI)
この改良された技術は、イェール大学とカロリンスカ研究所の研究者たちによって開発され、組織の発達、特定の疾患の理解、そしてその治療法について新たな光を投げかけることができる可能性があります。
本研究は、マウスの脳と人間の脳組織に技術を適用し、エピゲノムとトランスクリプトームを空間的にマッピングすることに成功しました。
この新しい技術は、組織内の精密な位置での遺伝子制御に対する洞察をもたらすことができます。また、個々の患者における癌を促進する遺伝子や腫瘍抑制遺伝子のエピジェネティックなメカニズムを理解する上で、将来的に個別化医療の進歩につながることが期待されています。
<関連情報>
- https://news.ki.se/new-technology-maps-where-and-how-cells-read-their-genome
- https://seas.yale.edu/news-events/news/new-technology-simultaneously-maps-gene-activity-and-expression
- https://www.nature.com/articles/s41586-023-05795-1
哺乳類組織の空間的エピゲノム-トランスクリプトームコプロファイリング Spatial epigenome–transcriptome co-profiling of mammalian tissues
Di Zhang,Yanxiang Deng,Petra Kukanja,Eneritz Agirre,Marek Bartosovic,Mingze Dong,Cong Ma,Sai Ma,Graham Su,Shuozhen Bao,Yang Liu,Yang Xiao,Gorazd B. Rosoklija,Andrew J. Dwork,J. John Mann,Kam W. Leong,Maura Boldrini,Liya Wang,Maximilian Haeussler,Benjamin J. Raphael,Yuval Kluger,Gonçalo Castelo-Branco & Rong Fan
Nature Published:15 March 2023
DOI:https://doi.org/10.1038/s41586-023-05795-1
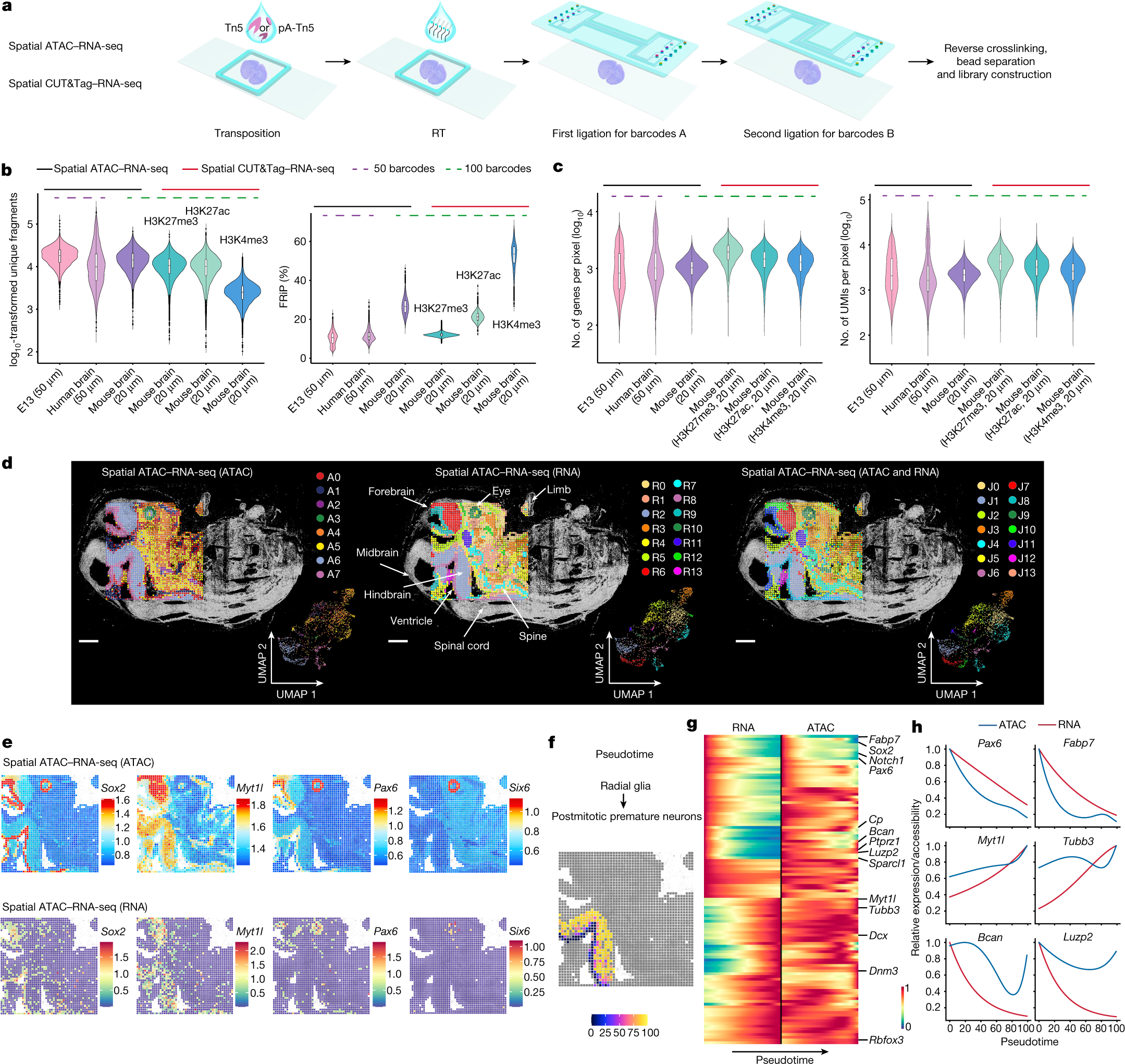
Abstract
Emerging spatial technologies, including spatial transcriptomics and spatial epigenomics, are becoming powerful tools for profiling of cellular states in the tissue context1,2,3,4,5. However, current methods capture only one layer of omics information at a time, precluding the possibility of examining the mechanistic relationship across the central dogma of molecular biology. Here, we present two technologies for spatially resolved, genome-wide, joint profiling of the epigenome and transcriptome by cosequencing chromatin accessibility and gene expression, or histone modifications (H3K27me3, H3K27ac or H3K4me3) and gene expression on the same tissue section at near-single-cell resolution. These were applied to embryonic and juvenile mouse brain, as well as adult human brain, to map how epigenetic mechanisms control transcriptional phenotype and cell dynamics in tissue. Although highly concordant tissue features were identified by either spatial epigenome or spatial transcriptome we also observed distinct patterns, suggesting their differential roles in defining cell states. Linking epigenome to transcriptome pixel by pixel allows the uncovering of new insights in spatial epigenetic priming, differentiation and gene regulation within the tissue architecture. These technologies are of great interest in life science and biomedical research.


