2023-05-02 スイス連邦工科大学ローザンヌ校(EPFL)
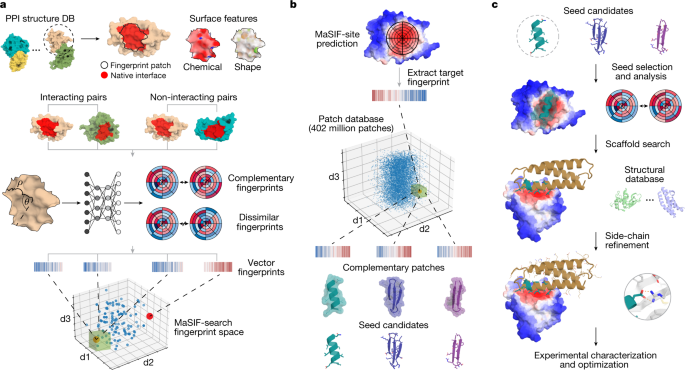
◆その研究チームは、表面化学的および幾何学的「指紋」に基づいて分子間の最適なマッチングを見つけることによって、計算的にタンパク質相互作用を設計することを目指していた。そして、この技術を用いて、新たなタンパク質結合体を設計することに成功し、Natureに論文が掲載された。
◆この技術は、がん治療や感染症治療などの分野で、計算上のタンパク質相互作用を制御することに大きな関心が寄せられている。
<関連情報>
- https://actu.epfl.ch/news/engineering-molecular-interactions-with-machine-le/
- https://www.nature.com/articles/s41586-023-05993-x
学習された表面指紋を用いたタンパク質相互作用のデノボデザイン De novo design of protein interactions with learned surface fingerprints
Pablo Gainza,Sarah Wehrle,Alexandra Van Hall-Beauvais,Anthony Marchand,Andreas Scheck,Zander Harteveld,Stephen Buckley,Dongchun Ni,Shuguang Tan,Freyr Sverrisson,Casper Goverde,Priscilla Turelli,Charlène Raclot,Alexandra Teslenko,Martin Pacesa,Stéphane Rosset,Sandrine Georgeon,Jane Marsden,Aaron Petruzzella,Kefang Liu,Zepeng Xu,Yan Chai,Pu Han,George F. Gao,Elisa Oricchio,Beat Fierz,Didier Trono,Henning Stahlberg,Michael Bronstein & Bruno E. Correia
Nature Published:26 April 2023
DOI:https://doi.org/10.1038/s41586-023-05993-x
Abstract
Physical interactions between proteins are essential for most biological processes governing life1. However, the molecular determinants of such interactions have been challenging to understand, even as genomic, proteomic and structural data increase. This knowledge gap has been a major obstacle for the comprehensive understanding of cellular protein–protein interaction networks and for the de novo design of protein binders that are crucial for synthetic biology and translational applications2,3,4,5,6,7,8,9. Here we use a geometric deep-learning framework operating on protein surfaces that generates fingerprints to describe geometric and chemical features that are critical to drive protein–protein interactions10. We hypothesized that these fingerprints capture the key aspects of molecular recognition that represent a new paradigm in the computational design of novel protein interactions. As a proof of principle, we computationally designed several de novo protein binders to engage four protein targets: SARS-CoV-2 spike, PD-1, PD-L1 and CTLA-4. Several designs were experimentally optimized, whereas others were generated purely in silico, reaching nanomolar affinity with structural and mutational characterization showing highly accurate predictions. Overall, our surface-centric approach captures the physical and chemical determinants of molecular recognition, enabling an approach for the de novo design of protein interactions and, more broadly, of artificial proteins with function.