2024-06-19 スイス連邦工科大学ローザンヌ校(EPFL)
<関連情報>
- https://actu.epfl.ch/news/mapping-the-biology-of-spinal-cord-injury-in-unpre/
- https://www.nature.com/articles/s41586-024-07504-y
Tabulae Paralyticaに掲載された脊髄損傷の単一細胞および空間アトラス Single-cell and spatial atlases of spinal cord injury in the Tabulae Paralytica
Michael A. Skinnider,Matthieu Gautier,Alan Yue Yang Teo,Claudia Kathe,Thomas H. Hutson,Achilleas Laskaratos,Alexandra de Coucy,Nicola Regazzi,Viviana Aureli,Nicholas D. James,Bernard Schneider,Michael V. Sofroniew,Quentin Barraud,Jocelyne Bloch,Mark A. Anderson,Jordan W. Squair & Grégoire Courtine
Nature Published:19 June 2024
DOI:https://doi.org/10.1038/s41586-024-07504-y
Abstract
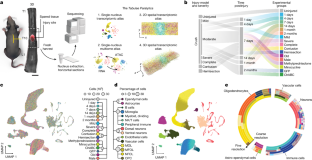
Here, we introduce the Tabulae Paralytica—a compilation of four atlases of spinal cord injury (SCI) comprising a single-nucleus transcriptome atlas of half a million cells, a multiome atlas pairing transcriptomic and epigenomic measurements within the same nuclei, and two spatial transcriptomic atlases of the injured spinal cord spanning four spatial and temporal dimensions. We integrated these atlases into a common framework to dissect the molecular logic that governs the responses to injury within the spinal cord1. The Tabulae Paralytica uncovered new biological principles that dictate the consequences of SCI, including conserved and divergent neuronal responses to injury; the priming of specific neuronal subpopulations to upregulate circuit-reorganizing programs after injury; an inverse relationship between neuronal stress responses and the activation of circuit reorganization programs; the necessity of re-establishing a tripartite neuroprotective barrier between immune-privileged and extra-neural environments after SCI and a failure to form this barrier in old mice. We leveraged the Tabulae Paralytica to develop a rejuvenative gene therapy that re-established this tripartite barrier, and restored the natural recovery of walking after paralysis in old mice. The Tabulae Paralytica provides a window into the pathobiology of SCI, while establishing a framework for integrating multimodal, genome-scale measurements in four dimensions to study biology and medicine.