2025-03-01 中国科学院 (CAS)
 Near-complete assembly of the Chinese Spring genome. (Image by IGDB)
Near-complete assembly of the Chinese Spring genome. (Image by IGDB)
<関連情報>
- https://english.cas.cn/newsroom/research_news/life/202503/t20250304_903021.shtml
- https://www.cell.com/molecular-plant/abstract/S1674-2052(25)00068-1
コムギChinese Springゲノムのほぼ完全なアセンブリと包括的アノテーション Near-complete assembly and comprehensive annotation of the wheat Chinese Spring genome
Zijian Wang∙ Lingfeng Miao∙ Kaiwen Tan∙ … ∙ Xiangdong Fu∙ Qixin Sun∙ Jian Chen
Molecular Plant Published:February 12, 2025
DOI:https://doi.org/10.1016/j.molp.2025.02.002
Abstract
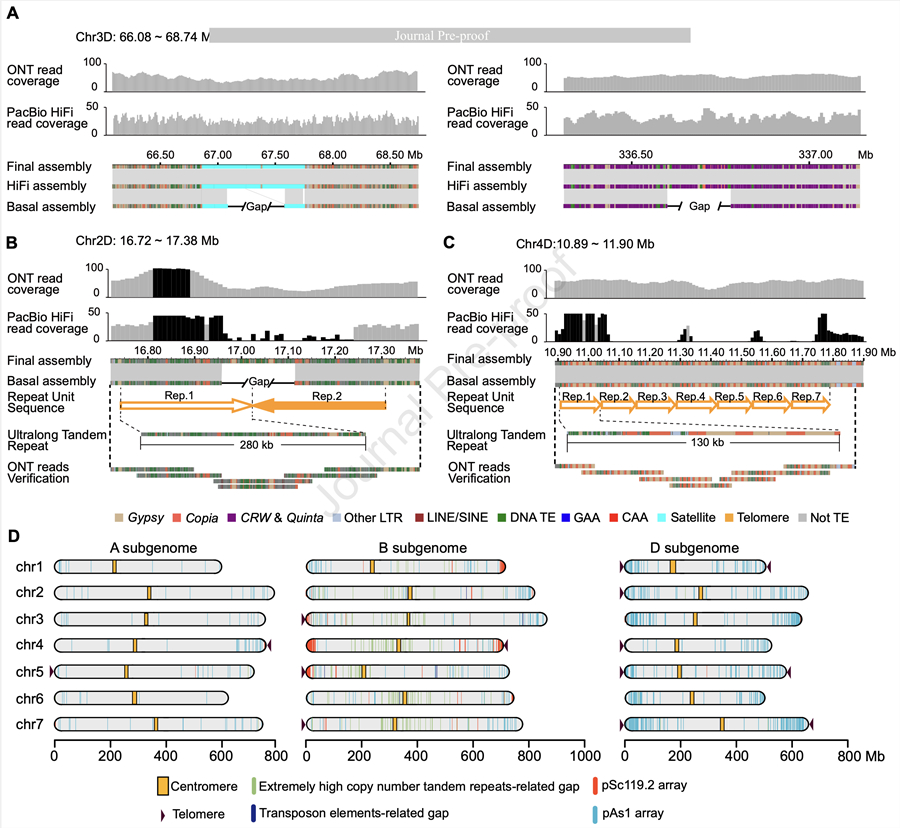
A complete reference genome assembly is crucial for biological research and genetic improvement. Owing to its large size and highly repetitive nature, there are numerous gaps in the globally used wheat Chinese Spring (CS) genome assembly. In this study, we generated a 14.46 Gb near-complete assembly of the CS genome, with a contig N50 of over 266 Mb and an overall base accuracy of 99.9963%. Among the 290 gaps that remained (26, 257, and 7 gaps from the A, B, and D subgenomes, respectively), 278 were extremely high-copy tandem repeats, whereas the remaining 12 were transposable-element-associated gaps. Four chromosome assemblies were completely gap-free, including chr1D, chr3D, chr4D, and chr5D. Extensive annotation of the near-complete genome revealed 151 405 high-confidence genes, of which 59 180 were newly annotated, including 7602 newly assembled genes. Except for the centromere of chr1B, which has a gap associated with superlong GAA repeat arrays, the centromeric sequences of all of the remaining 20 chromosomes were completely assembled. Our near-complete assembly revealed that the extent of tandem repeats, such as simple-sequence repeats, was highly uneven among different subgenomes. Similarly, the repeat compositions of the centromeres also varied among the three subgenomes. With the genome sequences of all six types of seed storage proteins (SSPs) fully assembled, the expression of ω-gliadin was found to be contributed entirely by the B subgenome, whereas the expression of the other five types of SSPs was most abundant from the D subgenome. The near-complete CS genome will serve as a valuable resource for genomic and functional genomic research and breeding of wheat as well as its related species.