2025-04-02 チャルマース工科大学
<関連情報>
- https://news.cision.com/chalmers/r/multi-resistance-in-bacteria-predicted-by-ai-model,c4127916
- https://www.nature.com/articles/s41467-025-57825-3
抗生物質耐性遺伝子の拡散を促進する遺伝的適合性と生態学的連結性 Genetic compatibility and ecological connectivity drive the dissemination of antibiotic resistance genes
David Lund,Marcos Parras-Moltó,Juan S. Inda-Díaz,Stefan Ebmeyer,D. G. Joakim Larsson,Anna Johnning & Erik Kristiansson
Nature Communications Published:16 March 2025
DOI:https://doi.org/10.1038/s41467-025-57825-3
Abstract
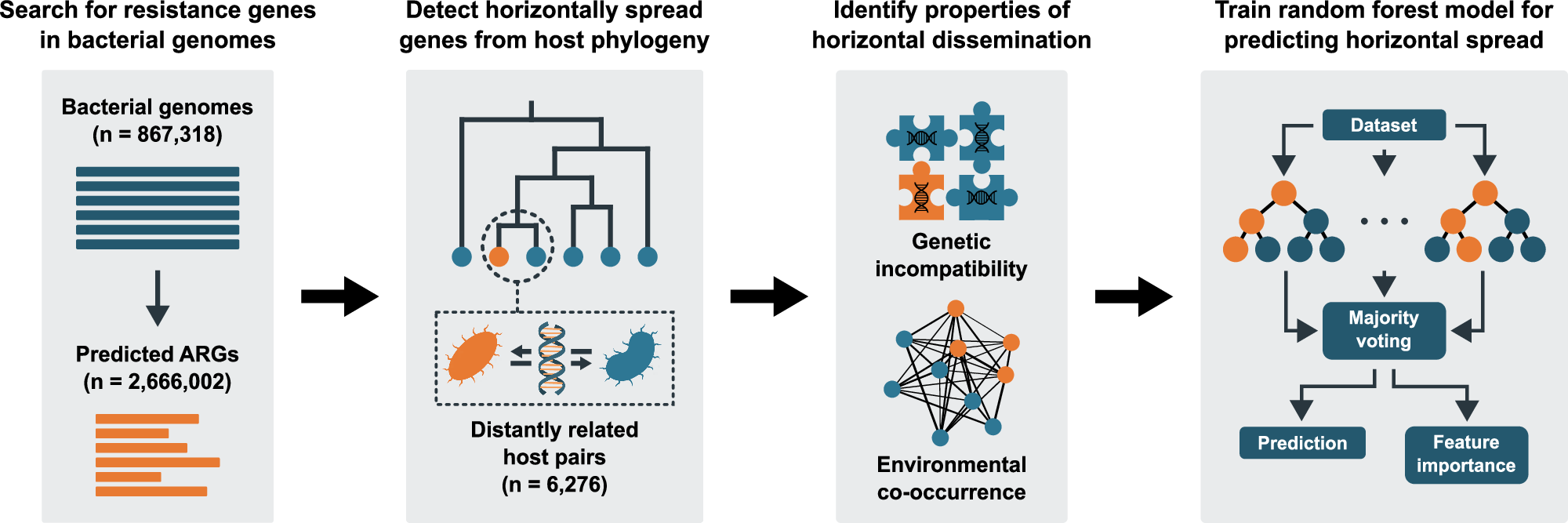
The dissemination of mobile antibiotic resistance genes (ARGs) via horizontal gene transfer is a significant threat to public health globally. The flow of ARGs into and between pathogens, however, remains poorly understood, limiting our ability to develop strategies for managing the antibiotic resistance crisis. Therefore, we aim to identify genetic and ecological factors that are fundamental for successful horizontal ARG transfer. We used a phylogenetic method to identify instances of horizontal ARG transfer in ~1 million bacterial genomes. This data was then integrated with >20,000 metagenomes representing animal, human, soil, water, and wastewater microbiomes to develop random forest models that can reliably predict horizontal ARG transfer between bacteria. Our results suggest that genetic incompatibility, measured as nucleotide composition dissimilarity, negatively influences the likelihood of transfer of ARGs between evolutionarily divergent bacteria. Conversely, environmental co-occurrence increases the likelihood, especially in humans and wastewater, in which several environment-specific dissemination patterns are observed. This study provides data-driven ways to predict the spread of ARGs and provides insights into the mechanisms governing this evolutionary process.