2025-04-29 カリフォルニア大学ロサンゼルス校(UCLA)
Jose Soto, Bhaduri Lab
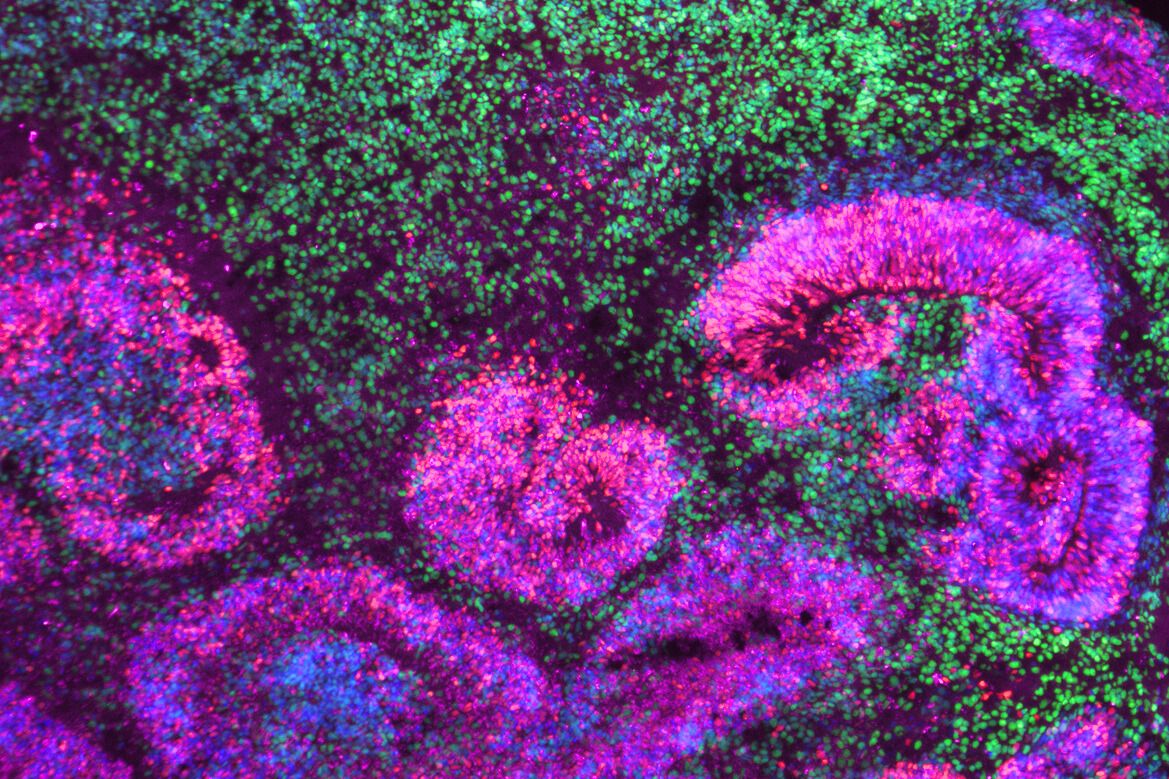
Microscopy image of a cortical organoid, a stem cell-derived model that mimics the developing human brain. The circular rosettes are clusters of neural stem cells giving rise to neurons, shown in green.
<関連情報>
- https://newsroom.ucla.edu/stories/crowdsourcing-brain-meta-atlas-reveals-new-development-disease-clues
- https://www.nature.com/articles/s41593-025-01933-2
分子アトラスの統合的解析により、ヒト大脳皮質における発生細胞サブタイプ指定を駆動するモジュールが明らかになった Integrated analysis of molecular atlases unveils modules driving developmental cell subtype specification in the human cortex
Patricia R. Nano,Elisa Fazzari,Daria Azizad,Antoni Martija,Claudia V. Nguyen,Sean Wang,Vanna Giang,Ryan L. Kan,Juyoun Yoo,Brittney Wick,Maximilian Haeussler & Aparna Bhaduri
Nature Neuroscience Published:21 April 2025
DOI:https://doi.org/10.1038/s41593-025-01933-2
Abstract
Human brain development requires generating diverse cell types, a process explored by single-cell transcriptomics. Through parallel meta-analyses of the human cortex in development (seven datasets) and adulthood (16 datasets), we generated over 500 gene co-expression networks that can describe mechanisms of cortical development, centering on peak stages of neurogenesis. These meta-modules show dynamic cell subtype specificities throughout cortical development, with several developmental meta-modules displaying spatiotemporal expression patterns that allude to potential roles in cell fate specification. We validated the expression of these modules in primary human cortical tissues. These include meta-module 20, a module elevated in FEZF2+ deep layer neurons that includes TSHZ3, a transcription factor associated with neurodevelopmental disorders. Human cortical chimeroid experiments validated that both FEZF2 and TSHZ3 are required to drive module 20 activity and deep layer neuron specification but through distinct modalities. These studies demonstrate how meta-atlases can engender further mechanistic analyses of cortical fate specification.


