2025-07-28 大阪大学
<関連情報>
- https://www.med.osaka-u.ac.jp/activities/results/2025year/edahiro2025-7-30
- https://www.nature.com/articles/s41588-025-02266-3
多層オミクスデータから単一細胞解像度で状態依存性免疫特性を解明する Deciphering state-dependent immune features from multi-layer omics data at single-cell resolution
Ryuya Edahiro,Go Sato,Tatsuhiko Naito,Yuya Shirai,Ryunosuke Saiki,Kyuto Sonehara,Yoshihiko Tomofuji,Kenichi Yamamoto,Shinichi Namba,Noah Sasa,Genta Nagao,Qingbo S. Wang,Yugo Takahashi,Takanori Hasegawa,Toshihiro Kishikawa,Ken Suzuki,Yu-Chen Liu,Daisuke Motooka,Ayako Takuwa,Hiromu Tanaka,Shuhei Azekawa,Japan COVID-19 Task Force,Ho Namkoong,Ryuji Koike,… Yukinori Okada
Nature Genetics Published:28 July 2025
DOI:https://doi.org/10.1038/s41588-025-02266-3
Abstract
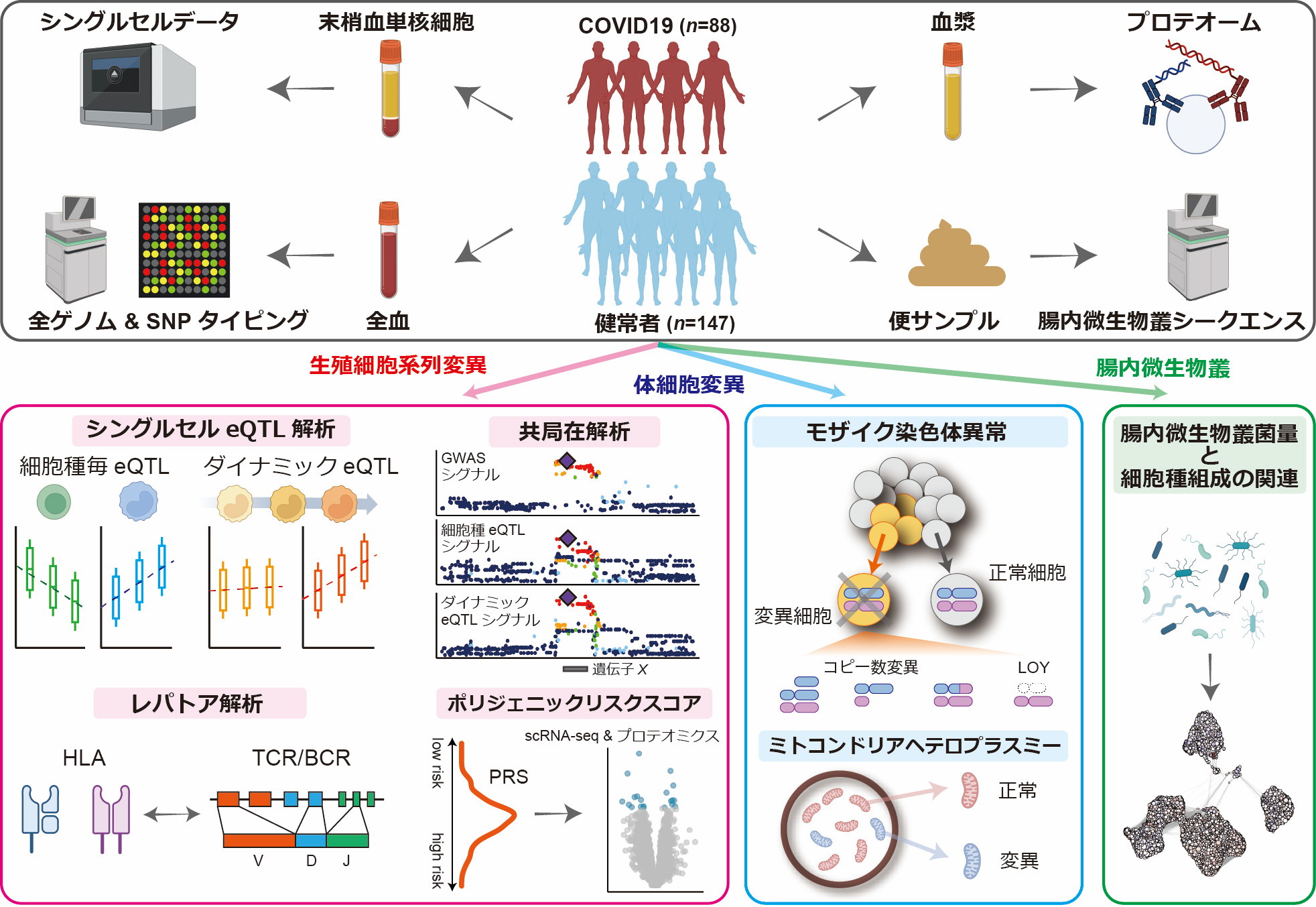
Current molecular quantitative trait locus catalogs are mostly at bulk resolution and centered on Europeans. Here, we constructed an immune cell atlas with single-cell transcriptomics of >1.5 million peripheral blood mononuclear cells, host genetics, plasma proteomics and gut metagenomics from 235 Japanese persons, including patients with coronavirus disease 2019 (COVID-19) and healthy individuals. We mapped germline genetic effects on gene expression within immune cell types and across cell states. We elucidated cell type- and context-specific human leukocyte antigen (HLA) and genome-wide associations with T and B cell receptor repertoires. Colocalization using dynamic genetic regulation provided better understanding of genome-wide association signals. Differential gene and protein expression analyses depicted cell type- and context-specific effects of polygenic risks. Various somatic mutations including mosaic chromosomal alterations, loss of Y chromosome and mitochondrial DNA (mtDNA) heteroplasmy were projected into single-cell resolution. We identified immune features specific to somatically mutated cells. Overall, immune cells are dynamically regulated in a cell state-dependent manner characterized with multiomic profiles.