2025-11-12 テキサスA&M大学
<関連情報>
- https://stories.tamu.edu/news/2025/11/12/texas-am-researchers-use-ai-to-identify-genetic-time-capsule-that-distinguishes-species/
- https://www.nature.com/articles/s41586-025-09740-2
古代の再結合砂漠は胎盤哺乳類の種分化超遺伝子である An ancient recombination desert is a speciation supergene in placental mammals
Nicole M. Foley,Richard G. Rasulis,Zoya Wani,Mayra N. Mendoza Cerna,Henrique V. Figueiró,Klaus Peter Koepfli,Terje Raudsepp & William J. Murphy
Nature Published:12 November 2025
DOI:https://doi.org/10.1038/s41586-025-09740-2
Abstract
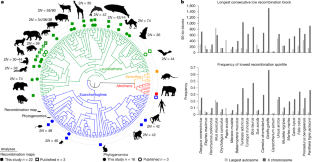
Gene flow between biological species is a common and often adaptive evolutionary phenomenon throughout the tree of life1,2,3,4. Given the pervasive nature of genetic exchange, a daunting challenge is how best to infer the correct relationships between species from the complex collection of histories arrayed across genomes5. The local rate of meiotic recombination influences the distribution of signatures of, and barriers to, gene flow during the early stages of speciation6. Still, a broader understanding of this relationship and its application to accurately discerning phylogeny is lacking due to a scarcity of recombination maps. Here we applied deep learning methods to genome alignments from 22 divergent placental mammal species to infer the evolution of the recombination landscape. We identified a large and evolutionarily conserved X-linked recombination desert constituting 30% of the chromosome. Recombination-aware phylogenomic analyses from 94 species revealed that the X-linked recombination desert is an ancient and recurrent barrier to gene flow and retains the species history when introgression dominates genome-wide ancestry. The functional basis for this supergene is manifold, enriched with genes that influence sex chromosome silencing and reproduction traits. Because the locus underpins reproductive isolation across ordinal lineages, it may represent a reliable marker for resolving challenging relationships across the mammalian phylogeny.