2025-12-18 カリフォルニア大学サンディエゴ校(UCSD)
<関連情報>
- https://today.ucsd.edu/story/could-your-genes-influence-the-gut-microbiome-of-others
- https://www.nature.com/articles/s41467-025-66105-z
実験用ラットの多コホート解析による宿主-マイクロバイオーム相互作用の遺伝的構造とメカニズム Genetic architecture and mechanisms of host-microbiome interactions from a multi-cohort analysis of outbred laboratory rats
Hélène Tonnelé,Denghui Chen,Felipe Morillo,Jorge Garcia-Calleja,Apurva S. Chitre,Benjamin B. Johnson,Thiago Missfeldt Sanches,Riyan Cheng,Marc Jan Bonder,Antonio Gonzalez,Tomasz Kosciolek,Anthony M. George,Wenyan Han,Katie Holl,Aidan Horvath,Keita Ishiwari,Christopher P. King,Alexander C. Lamparelli,Connor D. Martin,Angel Garcia Martinez,Alesa H. Netzley,Jordan A. Tripi,Tengfei Wang,Elena Bosch,… Amelie Baud
Nature Communications Published:18 December 2025
DOI:https://doi.org/10.1038/s41467-025-66105-z
Abstract
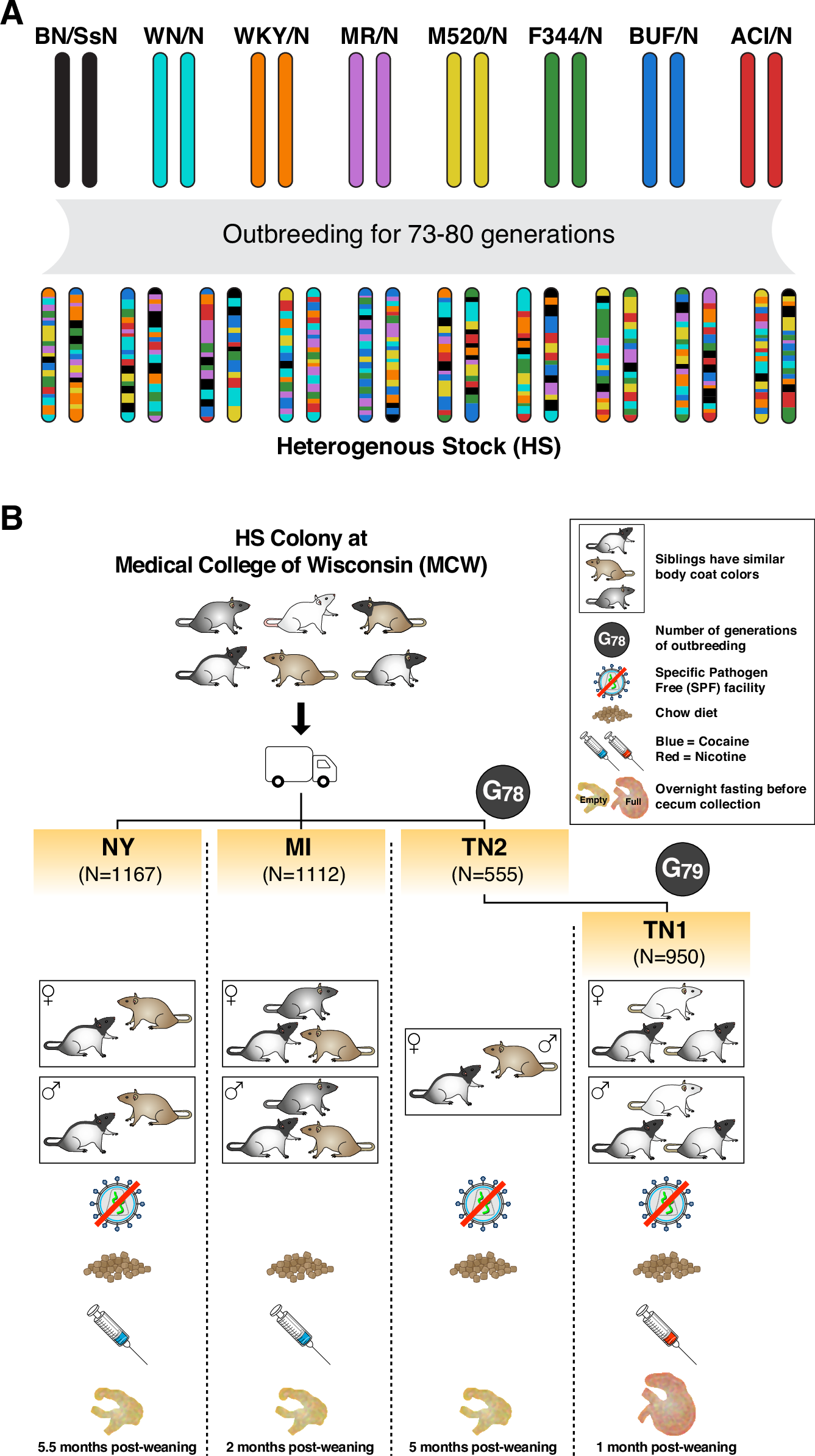
The intestinal microbiome influences health and disease. Its composition is affected by host genetics and environmental exposures. Understanding host genetic effects is critical but challenging in humans, due to the difficulty of detecting, mapping and interpreting them. To address this, we analyse host genetic effects in four cohorts of outbred laboratory rats exposed to distinct but controlled environments. We show that polygenic host genetic effects are consistent across cohort environments. We identify three replicated microbiome-associated loci, one of which involves the sialyltransferase gene St6galnac1 and Paraprevotella. We find a similar association in a human cohort, between ST6GAL1 and Paraprevotella, both of which have been linked with immune and infectious diseases. Moreover, we find indirect (i.e. social) genetic effects on microbiome phenotypes, which substantially increase the total genetic variance. Finally, we identify a novel mechanism whereby indirect genetic effects can contribute to “missing heritability”.