2026-02-03 東京科学大学
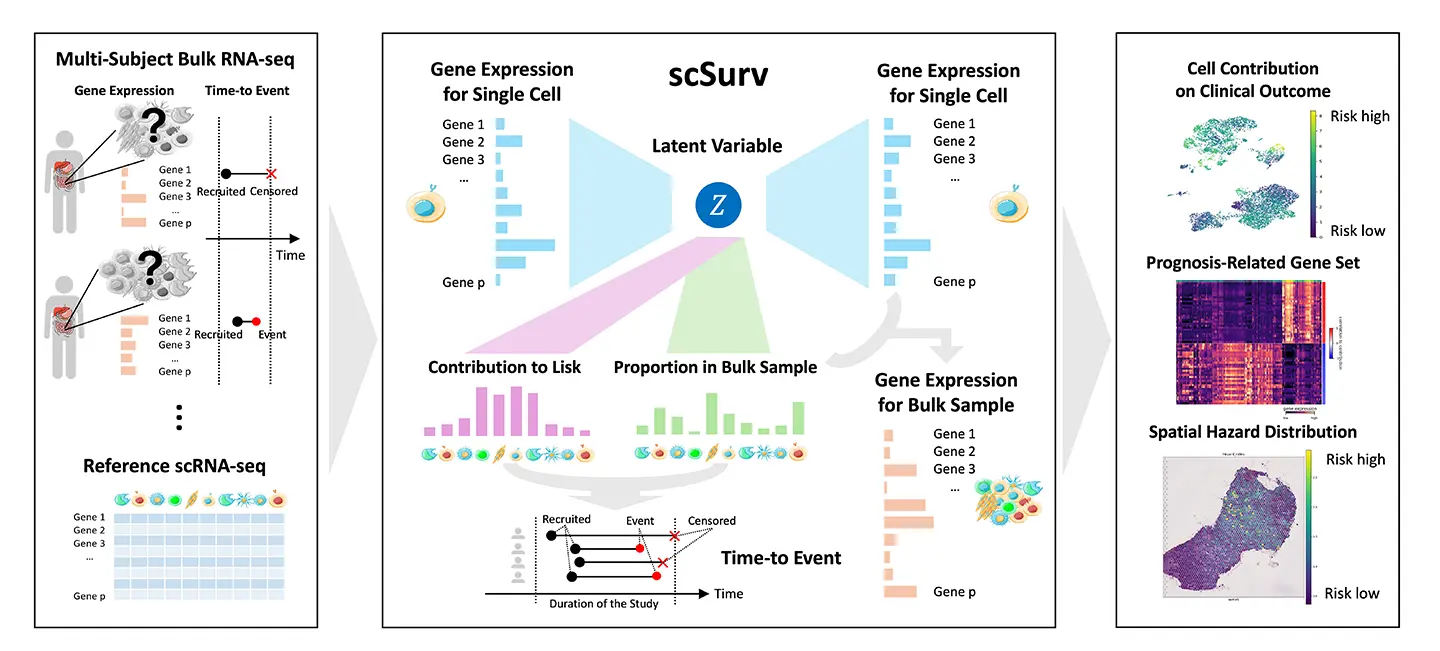
図1. scSurvの概念図。
<関連情報>
- https://www.isct.ac.jp/ja/news/ozwbdeconlq9
- https://www.isct.ac.jp/plugins/cms/component_download_file.php?type=2&pageId=&contentsId=1&contentsDataId=3168&prevId=&key=c662786a6f3f53f193f9e0f76993fa49.pdf
- https://academic.oup.com/bioinformatics/article/42/1/btaf671/8402136
scSurv:単一細胞生存解析のための深層生成モデル scSurv: a deep generative model for single-cell survival analysis
Chikara Mizukoshi ,Yasuhiro Kojima ,Shuto Hayashi ,Ko Abe ,Daisuke Kasugai ,Teppei Shimamura
Bioinformatics Published:22 December 2025
DOI:https://doi.org/10.1093/bioinformatics/btaf671
Abstract
Motivation
Single-cell omics analysis has unveiled the heterogeneity of various cell types within tumors. However, no methodology currently reveals how this heterogeneity influences cancer patient survival at single-cell resolution. Here, we introduce scSurv, combining a Cox proportional hazards model with a deep generative model of single-cell transcriptome, to estimate individual cellular contributions to clinical outcomes.
Results
The accuracy of scSurv was validated using both simulated and real datasets. This method identifies cells associated with favorable or adverse prognoses and extracts genes correlated with their contribution levels. In melanoma, scSurv reproduces known prognostic macrophage classifications and facilitates hazard mapping through spatial transcriptomics in renal cell carcinoma. We also identified genes consistently associated with prognosis across multiple cancers and demonstrated the applicability of this method to infectious diseases. scSurv is a novel framework for quantifying the heterogeneity of individual cellular effects on clinical outcomes.
Availability
The implementation of scSurv is available on GitHub (https://github.com/3254c/scSurv) and Zenodo (https://doi.org/10.5281/zenodo.17793054).