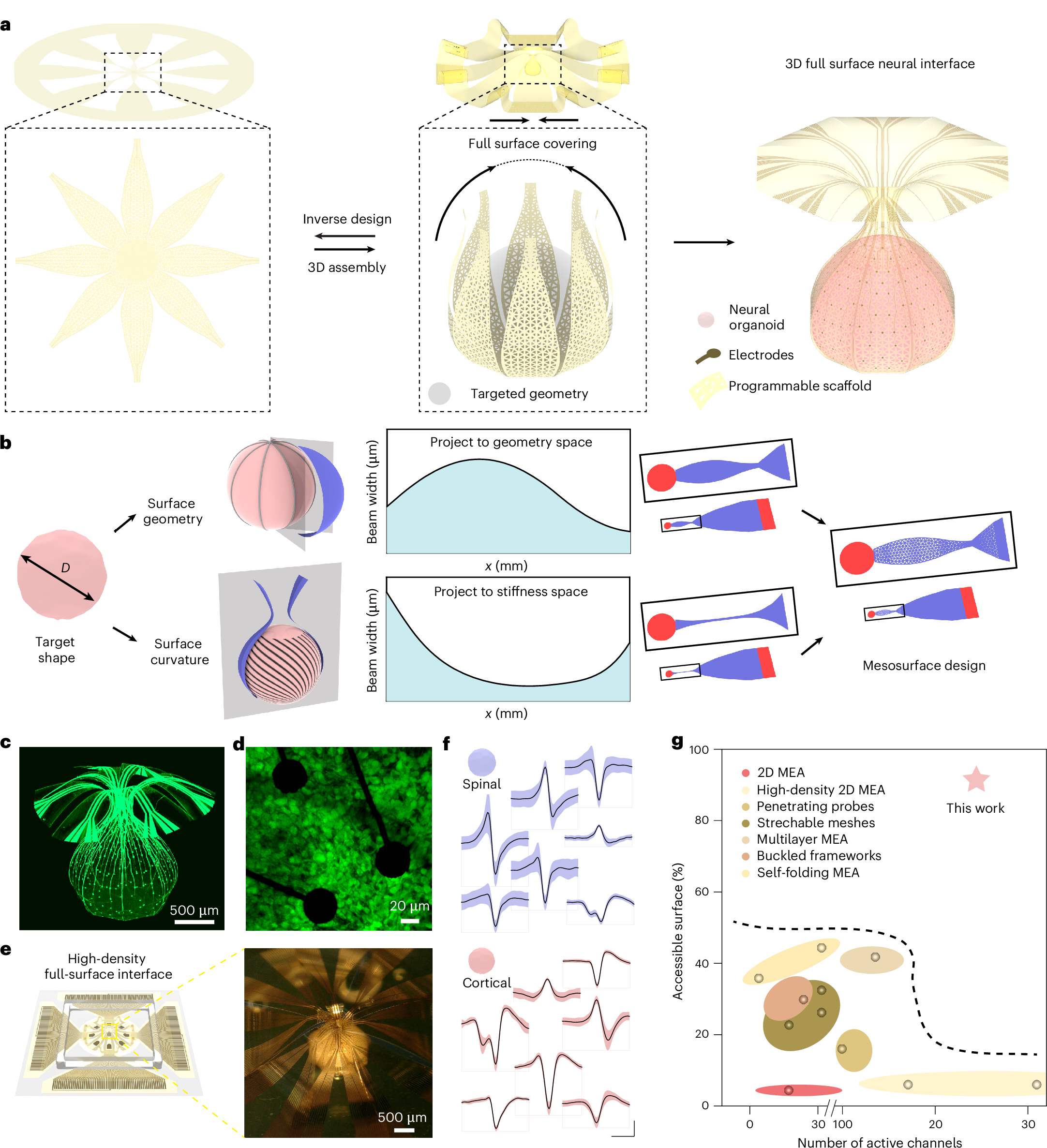
2026-02-16 マサチューセッツ工科大学(MIT)
Using a large language model (LLM), MIT researchers analyzed the genetic code of industrial yeast Komagataella phaffii — specifically, the codons that it uses. The new MIT model could then predict which codons would work best for manufacturing a given protein.Credits:Image: MIT News; iStock
<関連情報>
- https://news.mit.edu/2026/new-ai-model-could-cut-costs-developing-protein-drugs-0216
- https://www.pnas.org/doi/abs/10.1073/pnas.2522052123
Pichia-CLM: Komagataella phaffiiの言語モデルに基づくコドン最適化パイプライン Pichia-CLM: A language model–based codon optimization pipeline for Komagataella phaffii
Harini Narayanan and J. Christopher Love
Proceedings of the National Academy of Sciences Published:February 17, 2026
Significance
This paper presents Pichia–Codon language model (Pichia-CLM), a deep learning–based language model for codon optimization to enhance recombinant protein production in the industrially relevant host, Komagataella phaffii. Unlike conventional approaches that rely on codon usage bias metrics (CUB)—often providing a global score and ignoring sequence context—Pichia-CLM leverages the host genome to unbiasedly learn the amino acid-to-codon mapping. Prior deep learning models have attempted codon optimization but typically evaluated performance using CUB metrics with limited experimental validation. In contrast, we experimentally validate Pichia-CLM across six diverse protein classes of varying complexity and consistently observe superior expression titers compared to four commercial codon optimization tools. Our findings also reveal the limitations of using CUB metrics, showing a poor correlation between these scores and observed protein yields.
Abstract
The preference in synonymous codon usage—the so-called codon usage bias (CUB)—is governed by several factors such as the host organism, context and function of the gene, and the position of the codon within the gene itself. We demonstrated that this mapping can be learned from the host’s genome using language models and subsequently applied for codon optimization of heterologous proteins expressed by the host. This pipeline called Pichia–Codon language model (Pichia-CLM) was applied to the industrial host organism, Komagataella phaffii. With this approach, production of heterologous proteins was enhanced up to threefold compared to their native sequences. Furthermore, Pichia-CLM consistently yielded constructs with enhanced productivity for proteins of varied complexity, compared to commercially available tools. Finally, we showed that Pichia-CLM generates sequences resembling the properties of codon usage found in the host’s intrinsic host cell proteins and learned features such as avoiding negative cis-regulatory and repeat elements based on patterns in the genome data. These results show the potential of language models to unbiasedly learn patterns and design robust sequences for improved protein production.