タンパク質の相互作用を明らかにすることで、新薬の発見が可能になる Identifying protein interactions can help discover new medicines
2022-05-16 アメリカ国立研究所(PNNL)
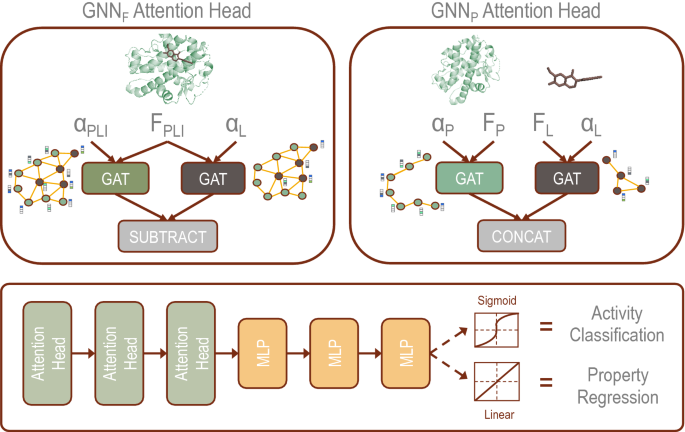
これらのGNNは、低分子化合物とタンパク質の相互作用を知るために、実験的に解像された薬剤候補分子と標的タンパク質両方の3次元原子構造を考慮する。この情報から、このモデルは、低分子化合物の標的タンパク質への結合親和性や生物物理学的特性(IC50など)を予測することができる。
<関連情報>
- https://www.pnnl.gov/news-media/decoding-protein-interactions-domain-aware-machine-learning
- https://www.nature.com/articles/s41598-022-10418-2
並列グラフ神経回路網を用いたタンパク質-リガンド相互作用の解読 Decoding the protein–ligand interactions using parallel graph neural networks
Carter Knutson,Mridula Bontha,Jenna A. Bilbrey & Neeraj Kumar
Scientific Reports Published:10 May 2022
DOI:https://doi.org/10.1038/s41598-022-10418-2
Abstract
Protein–ligand interactions (PLIs) are essential for biochemical functionality and their identification is crucial for estimating biophysical properties for rational therapeutic design. Currently, experimental characterization of these properties is the most accurate method, however, this is very time-consuming and labor-intensive. A number of computational methods have been developed in this context but most of the existing PLI prediction heavily depends on 2D protein sequence data. Here, we present a novel parallel graph neural network (GNN) to integrate knowledge representation and reasoning for PLI prediction to perform deep learning guided by expert knowledge and informed by 3D structural data. We develop two distinct GNN architectures is the base implementation that employs distinct featurization to enhance domain-awareness, while GNNP is a novel implementation that can predict with no prior knowledge of the intermolecular interactions. The comprehensive evaluation demonstrated that GNN can successfully capture the binary interactions between ligand and protein’s 3D structure with 0.979 test accuracy for GNNF and 0.958 for GNNP for predicting activity of a protein–ligand complex. These models are further adapted for regression tasks to predict experimental binding affinities and pIC50 crucial for compound’s potency and efficacy. We achieve a Pearson correlation coefficient of 0.66 and 0.65 on experimental affinity and 0.50 and 0.51 on pIC50 with GNNF and GNNP, respectively, outperforming similar 2D sequence based models. Our method can serve as an interpretable and explainable artificial intelligence (AI) tool for predicted activity, potency, and biophysical properties of lead candidates. To this end, we show the utility of GNNP on SARS-Cov-2 protein targets by screening a large compound library and comparing the prediction with the experimentally measured data.