アルツハイマー病の発症リスクを高める2つの新しい遺伝子が研究者によって発見されました。 Two new genes that raise a person’s risk of developing Alzheimer’s Disease have been discovered by researchers.
2022-11-21 カーディフ大学
カーディフ大学認知症研究所の所長であり、本研究の共著者であるジュリー・ウィリアムズ教授は、次のように述べています。「これらの知見は、脳の免疫システムやコレステロールの処理方法の違いなど、脳における非常に特異的な処理方法を指し示しています。これらの違いは、脳の機能に影響を与え、アルツハイマー病の発症につながる。また、これらの遺伝子の発見により、アルツハイマー病の基礎となるメカニズムを理解し、将来、新しい薬剤を用いた治療や遺伝子治療などの標的治療法を開発するための遺伝子モデルの構築が可能になります。」
本研究は、「Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer’s disease」と題し、Amsterdam UMCが主導し、Nature Geneticsに発表されました。
<関連情報>
- https://www.cardiff.ac.uk/news/view/2686031-new-alzheimers-genes-discovered-in-worlds-largest-study
- https://www.nature.com/articles/s41588-022-01208-7
エクソーム解析により、アルツハイマー病のリスク因子としてATP8B4とABCA1の稀少な損傷変異を同定 Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer’s disease
Henne Holstege,Marc Hulsman,Camille Charbonnier,Benjamin Grenier-Boley,Olivier Quenez,Detelina Grozeva,Jeroen G. J. van Rooij,Rebecca Sims,Shahzad Ahmad,Najaf Amin,Penny J. Norsworthy,Oriol Dols-Icardo,Holger Hummerich,Amit Kawalia,Philippe Amouyel,Gary W. Beecham,Claudine Berr,Joshua C. Bis,Anne Boland,Paola Bossù,Femke Bouwman,Jose Bras,Dominique Campion,J. Nicholas Cochran,Antonio Daniele,Jean-François Dartigues,Stéphanie Debette,Jean-François Deleuze,Nicola Denning,Anita L. DeStefano,Lindsay A. Farrer,Maria Victoria Fernández,Nick C. Fox,Daniela Galimberti,Emmanuelle Genin,Johan J. P. Gille,Yann Le Guen,Rita Guerreiro,Jonathan L. Haines,Clive Holmes,M. Arfan Ikram,M. Kamran Ikram,Iris E. Jansen,Robert Kraaij,Marc Lathrop,Afina W. Lemstra,Alberto Lleó,Lauren Luckcuck,Marcel M. A. M. Mannens,Rachel Marshall,Eden R. Martin,Carlo Masullo,Richard Mayeux,Patrizia Mecocci,Alun Meggy,Merel O. Mol,Kevin Morgan,Richard M. Myers,Benedetta Nacmias,Adam C. Naj,Valerio Napolioni,Florence Pasquier,Pau Pastor,Margaret A. Pericak-Vance,Rachel Raybould,Richard Redon,Marcel J. T. Reinders,Anne-Claire Richard,Steffi G. Riedel-Heller,Fernando Rivadeneira,Stéphane Rousseau,Natalie S. Ryan,Salha Saad,Pascual Sanchez-Juan,Gerard D. Schellenberg,Philip Scheltens,Jonathan M. Schott,Davide Seripa,Sudha Seshadri,Daoud Sie,Erik A. Sistermans,Sandro Sorbi,Resie van Spaendonk,Gianfranco Spalletta,Niccolo’ Tesi,Betty Tijms,André G. Uitterlinden,Sven J. van der Lee,Pieter Jelle Visser,Michael Wagner,David Wallon,Li-San Wang,Aline Zarea,Jordi Clarimon,John C. van Swieten,Michael D. Greicius,Jennifer S. Yokoyama,Carlos Cruchaga,John Hardy,Alfredo Ramirez,Simon Mead,Wiesje M. van der Flier,Cornelia M. van Duijn,Julie Williams,Gaël Nicolas,Céline Bellenguez & Jean-Charles Lambert
Nature Genetics Published21 November 2022
DOIhttps://doi.org/10.1038/s41588-022-01208-7
Abstract
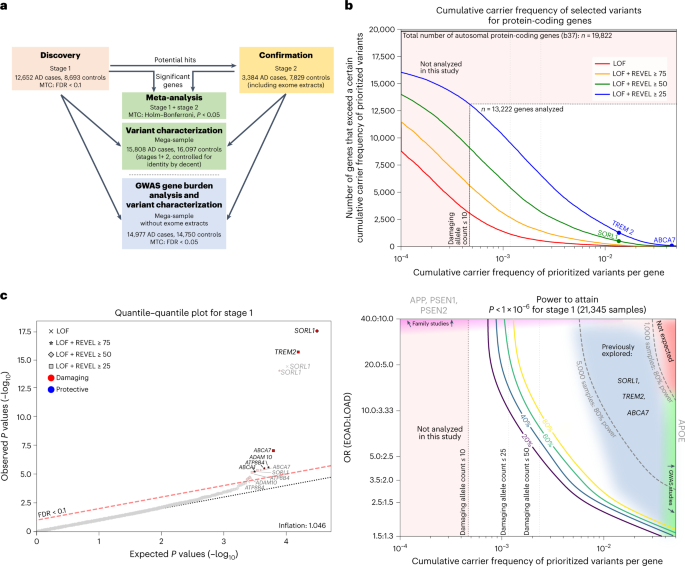
Alzheimer’s disease (AD), the leading cause of dementia, has an estimated heritability of approximately 70%1. The genetic component of AD has been mainly assessed using genome-wide association studies, which do not capture the risk contributed by rare variants2. Here, we compared the gene-based burden of rare damaging variants in exome sequencing data from 32,558 individuals—16,036 AD cases and 16,522 controls. Next to variants in TREM2, SORL1 and ABCA7, we observed a significant association of rare, predicted damaging variants in ATP8B4 and ABCA1 with AD risk, and a suggestive signal in ADAM10. Additionally, the rare-variant burden in RIN3, CLU, ZCWPW1 and ACE highlighted these genes as potential drivers of respective AD-genome-wide association study loci. Variants associated with the strongest effect on AD risk, in particular loss-of-function variants, are enriched in early-onset AD cases. Our results provide additional evidence for a major role for amyloid-β precursor protein processing, amyloid-β aggregation, lipid metabolism and microglial function in AD.