2023-03-01 カリフォルニア大学サンタバーバラ校(UCSB)
研究は、免疫と代謝に関連する特性が淘汰に関与していることを示している。
研究チームは、これらの熱帯集団が多くの寄生虫や様々な病原体にさらされている一方、ツチマネはめったに心血管疾患や認知症に苦しまないことが先行研究で示されていることを考慮に入れた。
これにより、ツチマネのゲノムが免疫と代謝に関連する特性に対して淘汰を受けたことが示唆された。
また、先住民族のDNAを研究するためには、公正さ、平等さ、および先住民族のデータ主権の必要性を考慮した、新しいデータ共有システムを開発することが重要であると強調されている。
<関連情報>
- https://www.news.ucsb.edu/2023/020860/special-selection
- https://www.pnas.org/doi/10.1073/pnas.2207544120
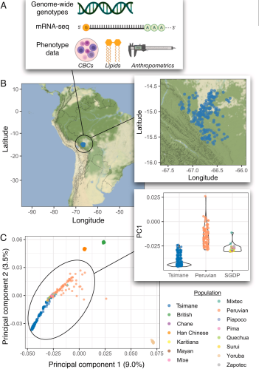
ボリビアの低地2集団における健康に関連する免疫・代謝遺伝子の自然選択 Natural selection of immune and metabolic genes associated with health in two lowland Bolivian populations
Amanda J. Lea,Angela Garcia,Jesusa Arevalo,Julien F. Ayroles ,Kenneth Buetow,Steve W. Cole,Daniel Eid Rodriguez ,Maguin Gutierrez,Heather M. Highland,Paul L. Hooper,Anne Justice,Thomas Kraft,Kari E. North,Jonathan Stieglitz,Hillard Kaplan,Benjamin C. Trumble,Michael D. Gurven
Proceedings of the National Academy of Sciences Published:December 27, 2022
DOI:https://doi.org/10.1073/pnas.2207544120
Significance
Humans have adapted to diverse environments, but few studies have linked regions of the genome with evidence of selection to phenotypes. We generated an integrative dataset of genomic, transcriptomic, and biomarker data for the Tsimane and the Moseten—two indigenous Amerindian populations in Bolivia. From analyses focused on Tsimane individuals, we found evidence for adaptation at genes and traits involved in immunity, which makes sense given that infection has strong effects on physiology and fitness in this population. Using phenotypic data from both populations, we were able to link genotypes to the immune and metabolic traits that are potentially advantageous. This study expands our knowledge of natural selection in Amerindians and uncovers previously undescribed loci of evolutionary and biomedical relevance.
Abstract
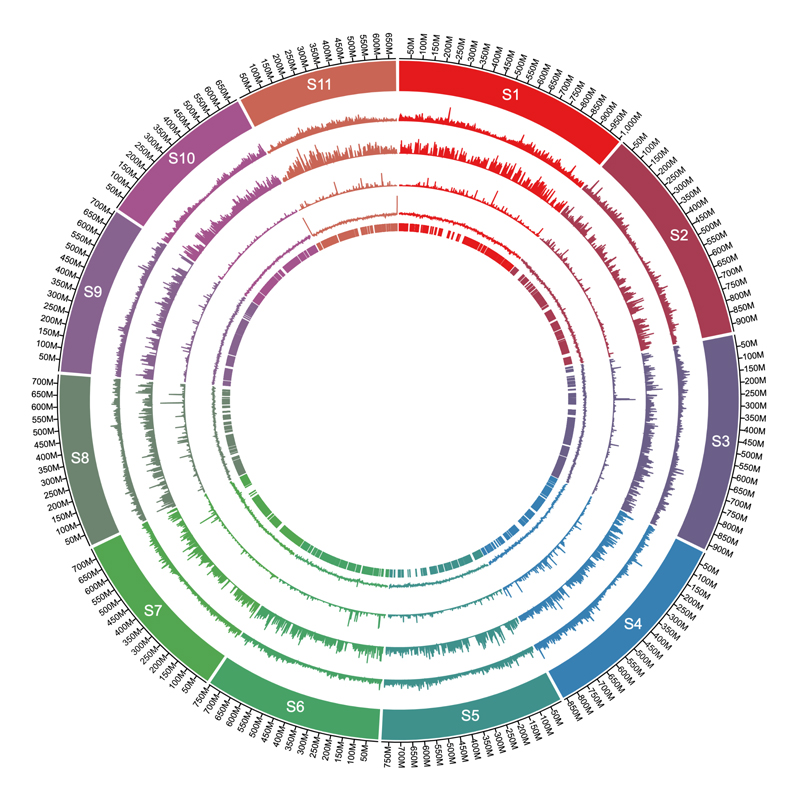
A growing body of work has addressed human adaptations to diverse environments using genomic data, but few studies have connected putatively selected alleles to phenotypes, much less among underrepresented populations such as Amerindians. Studies of natural selection and genotype–phenotype relationships in underrepresented populations hold potential to uncover previously undescribed loci underlying evolutionarily and biomedically relevant traits. Here, we worked with the Tsimane and the Moseten, two Amerindian populations inhabiting the Bolivian lowlands. We focused most intensively on the Tsimane, because long-term anthropological work with this group has shown that they have a high burden of both macro and microparasites, as well as minimal cardiometabolic disease or dementia. We therefore generated genome-wide genotype data for Tsimane individuals to study natural selection, and paired this with blood mRNA-seq as well as cardiometabolic and immune biomarker data generated from a larger sample that included both populations. In the Tsimane, we identified 21 regions that are candidates for selective sweeps, as well as 5 immune traits that show evidence for polygenic selection (e.g., C-reactive protein levels and the response to coronaviruses). Genes overlapping candidate regions were strongly enriched for known involvement in immune-related traits, such as abundance of lymphocytes and eosinophils. Importantly, we were also able to draw on extensive phenotype information for the Tsimane and Moseten and link five regions (containing PSD4, MUC21 and MUC22, TOX2, ANXA6, and ABCA1) with biomarkers of immune and metabolic function. Together, our work highlights the utility of pairing evolutionary analyses with anthropological and biomedical data to gain insight into the genetic basis of health-related traits.