腫瘍抑制因子が破綻する仕組みを解明することで、新たながん治療法につながる可能性がある Understanding how tumor suppressors fail could lead to new cancer treatments
2023-03-01 ペンシルベニア州立大学(PennState)
この発見により、将来的に新しい治療法の開発が期待されています。
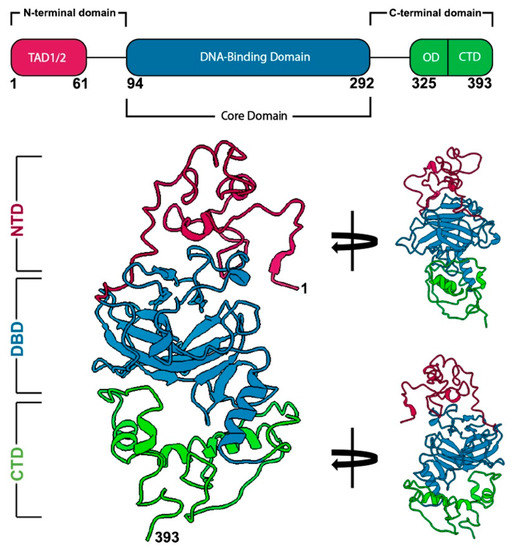
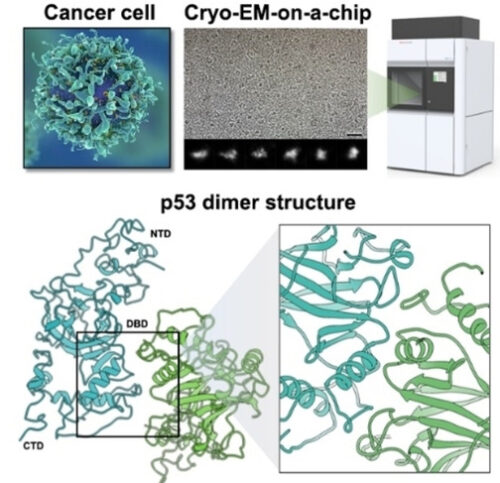
研究チームは、がん細胞から分離した個々のp53タンパク質を用いて、電子顕微鏡を用いたcryo-EM技術を使用して、p53の全長構造を解明しました。
この成果は、ChemBioChemおよびInternational Journal of Molecular Sciencesに掲載されました。
将来的には、この研究チームは、p53の突然変異が重要な役割を果たす膵臓および卵巣がんを研究する予定です
<関連情報>
- https://www.psu.edu/news/engineering/story/guarding-genome-researchers-uncover-full-3d-structure-p53-protein/
- https://www.mdpi.com/1422-0067/23/23/15267
- https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cbic.202200310
p53の完全なモデルで、ホットスポット変異の影響をより詳しく知ることができる Complete Models of p53 Better Inform the Impact of Hotspot Mutations
Maria J. Solares and Deborah F. Kelly
International Journal of Molecular Sciences Published: 3 December 2022
DOI:https://doi.org/10.3390/ijms232315267
Abstract
Mutations in tumor suppressor genes often lead to cancerous phenotypes. Current treatments leverage signaling pathways that are often compromised by disease-derived deficiencies in tumor suppressors. P53 falls into this category as genetic mutations lead to physical changes in the protein that impact multiple cellular pathways. Here, we show the first complete structural models of mutated p53 to reveal how hotspot mutations physically deviate from the wild-type protein. We employed a recently determined structure for the p53 monomer to map seven frequent clinical mutations using computational modeling approaches. Results showed that missense mutations often changed the conformational structure of p53 in the DNA-binding site along with its electrostatic surface charges. We posit these changes may amplify the toxic effects of these hotspot mutations by destabilizing an important zinc ion coordination region in p53 to impede proper DNA interactions. These results highlight the imperative need for new studies on patient-derived proteins that may assist in redesigning structure-informed targeted therapies.
マイクロプロセッサー材料を用いたヒトがんタンパク質の高解像度イメージング High-Resolution Imaging of Human Cancer Proteins Using Microprocessor Materials
Maria J. Solares, G. M. Jonaid, William Y. Luqiu, Samantha Berry, Janki Khadela, Yanping Liang, Madison C. Evans, Dr. Kevin J. Pridham, Dr. William J. Dearnaley, Prof. Dr. Zhi Sheng, Prof. Dr. Deborah F. Kelly
ChemBioChem Published: 05 July 2022
DOI:https://doi.org/10.1002/cbic.202200310
Abstract
Mutations in tumor suppressor genes, such as Tumor Protein 53 (TP53), are heavily implicated in aggressive cancers giving rise to gain- and loss-of-function phenotypes. While individual domains of the p53 protein have been studied extensively, structural information for full-length p53 remains incomplete. Functionalized microprocessor chips (microchips) with properties amenable to electron microscopy permitted us to visualize complete p53 assemblies for the first time. The new structures revealed p53 in an inactive dimeric state independent of DNA binding. Residues located at the protein-protein interface corresponded with modification sites in cancer-related hot spots. Changes in these regions may amplify the toxic effects of clinical mutations. Taken together, these results contribute advances in technology and imaging approaches to decode native protein models in different states of activation.