2023-07-19 カリフォルニア大学サンディエゴ校(UCSD)
◆研究者たちは、90人以上の患者を対象に、健康な腎臓と疾患を抱える腎臓の細胞状態をマッピングした、これまでに行われた最大の単一細胞アトラスを作成しました。このアトラスは、急性腎障害後の腎臓疾患の進行をよりよく理解するための基盤として利用される予定です。
◆急性腎障害は、腎臓が急激に血液から廃棄物をろ過する能力を失う状態です。研究者たちは、さまざまなタイプの細胞をアトラスに組み込むことで、疾患進行に寄与している可能性がある細胞タイプを特定しようとしました。その結果、腎臓の損傷細胞は、一部は修復をし、また一部は修復が不可能な状態に移行する要因が何であるかを知ることができました。
◆この洞察により、早期のカウンセリングがアルコールの回避につながる可能性があり、遺伝的原因がわかれば、より効果的な治療薬の開発も可能になるでしょう。
<関連情報>
- https://today.ucsd.edu/story/single-cell-atlas-of-the-human-kidney-provides-new-resources-to-study-kidney-disease
- https://www.nature.com/articles/s41586-023-05769-3
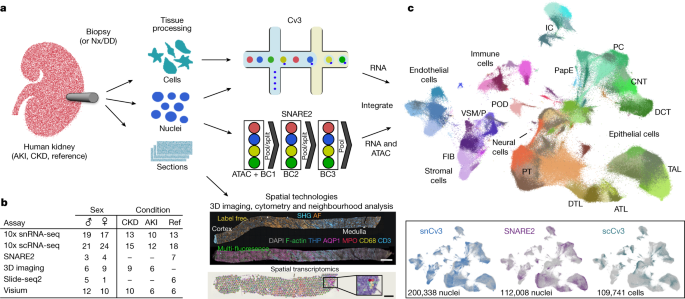
ヒト腎臓における健常細胞と傷害細胞の状態とニッチのアトラス An atlas of healthy and injured cell states and niches in the human kidney
Blue B. Lake,Rajasree Menon,Seth Winfree,Qiwen Hu,Ricardo Melo Ferreira,Kian Kalhor,Daria Barwinska,Edgar A. Otto,Michael Ferkowicz,Dinh Diep,Nongluk Plongthongkum,Amanda Knoten,Sarah Urata,Laura H. Mariani,Abhijit S. Naik,Sean Eddy,Bo Zhang,Yan Wu,Diane Salamon,James C. Williams,Xin Wang,Karol S. Balderrama,Paul J. Hoover,Evan Murray,Jamie L. Marshall,Teia Noel,Anitha Vijayan,Austin Hartman,Fei Chen,Sushrut S. Waikar,Sylvia E. Rosas,Francis P. Wilson,Paul M. Palevsky,Krzysztof Kiryluk,John R. Sedor,Robert D. Toto,Chirag R. Parikh,Eric H. Kim,Rahul Satija,Anna Greka,Evan Z. Macosko,Peter V. Kharchenko,Joseph P. Gaut,Jeffrey B. Hodgin,KPMP Consortium,Michael T. Eadon,Pierre C. Dagher,Tarek M. El-Achkar,Kun Zhang,Matthias Kretzler & Sanjay Jain
Nature Published:19 July 2023
DOI:https://doi.org/10.1038/s41586-023-05769-3
Abstract
Understanding kidney disease relies on defining the complexity of cell types and states, their associated molecular profiles and interactions within tissue neighbourhoods1. Here we applied multiple single-cell and single-nucleus assays (>400,000 nuclei or cells) and spatial imaging technologies to a broad spectrum of healthy reference kidneys (45 donors) and diseased kidneys (48 patients). This has provided a high-resolution cellular atlas of 51 main cell types, which include rare and previously undescribed cell populations. The multi-omic approach provides detailed transcriptomic profiles, regulatory factors and spatial localizations spanning the entire kidney. We also define 28 cellular states across nephron segments and interstitium that were altered in kidney injury, encompassing cycling, adaptive (successful or maladaptive repair), transitioning and degenerative states. Molecular signatures permitted the localization of these states within injury neighbourhoods using spatial transcriptomics, while large-scale 3D imaging analysis (around 1.2 million neighbourhoods) provided corresponding linkages to active immune responses. These analyses defined biological pathways that are relevant to injury time-course and niches, including signatures underlying epithelial repair that predicted maladaptive states associated with a decline in kidney function. This integrated multimodal spatial cell atlas of healthy and diseased human kidneys represents a comprehensive benchmark of cellular states, neighbourhoods, outcome-associated signatures and publicly available interactive visualizations.


