2023-11-15 マックス・プランク研究所
◆この手法は、健康な細胞でのタンパク質の相互作用を理解し、がんやハンチントン病などの疾患と関連する新しい接続を発見するために使用され、将来的な治療法の開発に寄与する可能性があります。酵母を用いた研究は、その細胞機能が人間との類似性からくるものであり、タンパク質が社会的に相互作用する様子を示しています。
<関連情報>
- https://www.mpg.de/21120471/1115-bioc-the-social-network-of-proteins-153945-x
- https://www.nature.com/articles/s41586-023-06739-5
酵母タンパク質インタラクトームの社会的・構造的構造 The social and structural architecture of the yeast protein interactome
André C. Michaelis,Andreas-David Brunner,Maximilian Zwiebel,Florian Meier,Maximilian T. Strauss,Isabell Bludau & Matthias Mann
Nature Published:15 November 2023
DOI:https://doi.org/10.1038/s41586-023-06739-5
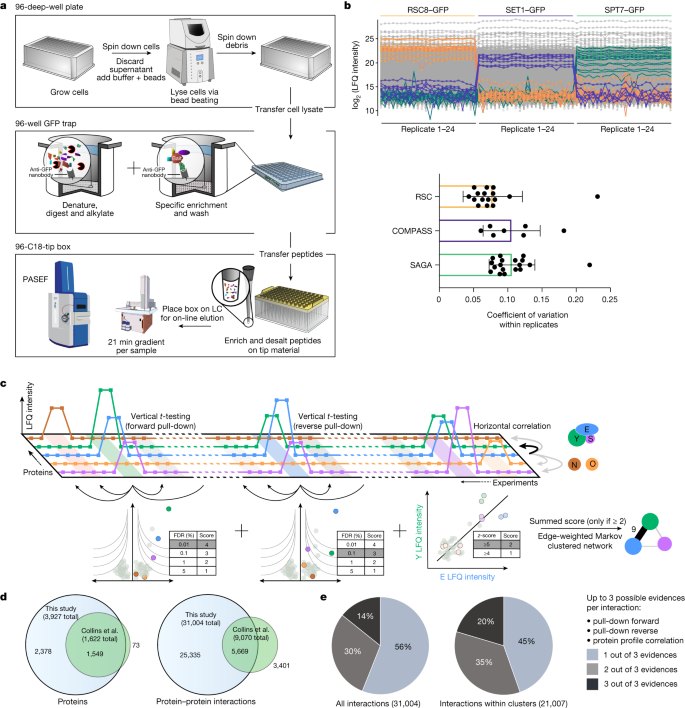
Abstract
Cellular functions are mediated by protein–protein interactions, and mapping the interactome provides fundamental insights into biological systems. Affinity purification coupled to mass spectrometry is an ideal tool for such mapping, but it has been difficult to identify low copy number complexes, membrane complexes and complexes that are disrupted by protein tagging. As a result, our current knowledge of the interactome is far from complete, and assessing the reliability of reported interactions is challenging. Here we develop a sensitive high-throughput method using highly reproducible affinity enrichment coupled to mass spectrometry combined with a quantitative two-dimensional analysis strategy to comprehensively map the interactome of Saccharomyces cerevisiae. Thousand-fold reduced volumes in 96-well format enabled replicate analysis of the endogenous GFP-tagged library covering the entire expressed yeast proteome1. The 4,159 pull-downs generated a highly structured network of 3,927 proteins connected by 31,004 interactions, doubling the number of proteins and tripling the number of reliable interactions compared with existing interactome maps2. This includes very-low-abundance epigenetic complexes, organellar membrane complexes and non-taggable complexes inferred by abundance correlation. This nearly saturated interactome reveals that the vast majority of yeast proteins are highly connected, with an average of 16 interactors. Similar to social networks between humans, the average shortest distance between proteins is 4.2 interactions. AlphaFold-Multimer provided novel insights into the functional roles of previously uncharacterized proteins in complexes. Our web portal (www.yeast-interactome.org) enables extensive exploration of the interactome dataset.