2024-05-08 カロリンスカ研究所(KI)
<関連情報>
- https://news.ki.se/new-technology-changes-how-proteins-in-individual-cells-are-studied
- https://www.nature.com/articles/s41592-024-02268-9
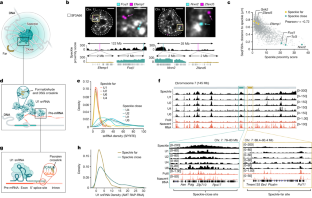
「モレキュラーピクセレーション」:配列決定による単一細胞の空間プロテオミクス Molecular pixelation: spatial proteomics of single cells by sequencing
Filip Karlsson,Tomasz Kallas,Divya Thiagarajan,Max Karlsson,Maud Schweitzer,Jose Fernandez Navarro,Louise Leijonancker,Sylvain Geny,Erik Pettersson,Jan Rhomberg-Kauert,Ludvig Larsson,Hanna van Ooijen,Stefan Petkov,Marcela González-Granillo,Jessica Bunz,Johan Dahlberg,Michele Simonetti,Prajakta Sathe,Petter Brodin,Alvaro Martinez Barrio & Simon Fredriksson
Nature Methods Published:08 May 2024
DOI:https://doi.org/10.1038/s41592-024-02268-9
Abstract
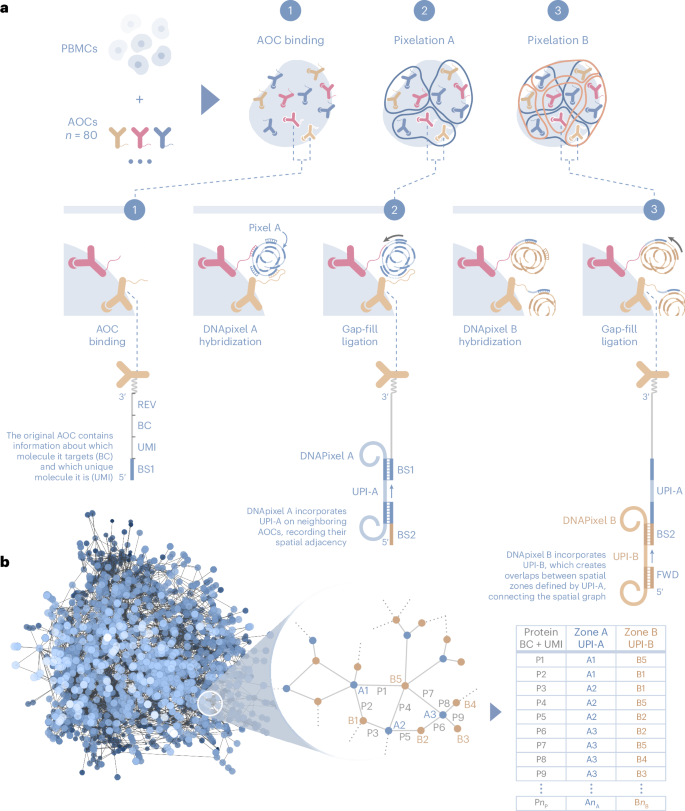
The spatial distribution of cell surface proteins governs vital processes of the immune system such as intercellular communication and mobility. However, fluorescence microscopy has limited scalability in the multiplexing and throughput needed to drive spatial proteomics discoveries at subcellular level. We present Molecular Pixelation (MPX), an optics-free, DNA sequence-based method for spatial proteomics of single cells using antibody–oligonucleotide conjugates (AOCs) and DNA-based, nanometer-sized molecular pixels. The relative locations of AOCs are inferred by sequentially associating them into local neighborhoods using the sequence-unique DNA pixels, forming >1,000 spatially connected zones per cell in 3D. For each single cell, DNA-sequencing reads are computationally arranged into spatial proteomics networks for 76 proteins. By studying immune cell dynamics using spatial statistics on graph representations of the data, we identify known and new patterns of spatial organization of proteins on chemokine-stimulated T cells, highlighting the potential of MPX in defining cell states by the spatial arrangement of proteins.