2024-05-15 ロスアラモス国立研究所(LANL)
<関連情報>
- https://discover.lanl.gov/news/0514-machine-learning/
- https://www.nature.com/articles/s42004-024-01161-y
緑膿菌の外膜を通過する化合物の分子記述子を用いた透過予測 Predicting permeation of compounds across the outer membrane of P. aeruginosa using molecular descriptors
Pedro D. Manrique,Inga V. Leus,César A. López,Jitender Mehla,Giuliano Malloci,Silvia Gervasoni,Attilio V. Vargiu,Rama K. Kinthada,Liam Herndon,Nicolas W. Hengartner,John K. Walker,Valentin V. Rybenkov,Paolo Ruggerone,Helen I. Zgurskaya & S. Gnanakaran
Communications Chemistry Published:12 April 2024
DOI:https://doi.org/10.1038/s42004-024-01161-y
Abstract
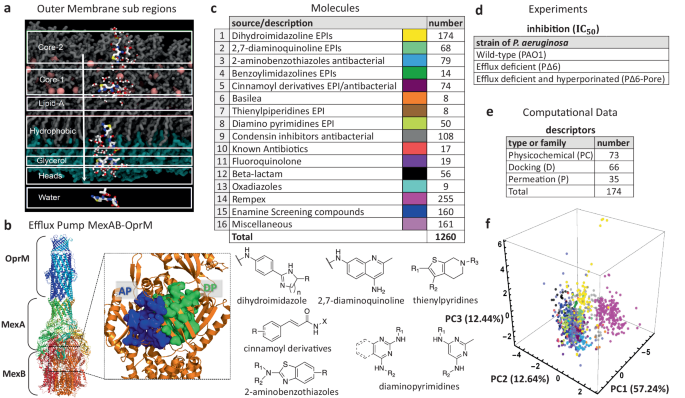
The ability Gram-negative pathogens have at adapting and protecting themselves against antibiotics has increasingly become a public health threat. Data-driven models identifying molecular properties that correlate with outer membrane (OM) permeation and growth inhibition while avoiding efflux could guide the discovery of novel classes of antibiotics. Here we evaluate 174 molecular descriptors in 1260 antimicrobial compounds and study their correlations with antibacterial activity in Gram-negative Pseudomonas aeruginosa. The descriptors are derived from traditional approaches quantifying the compounds’ intrinsic physicochemical properties, together with, bacterium-specific from ensemble docking of compounds targeting specific MexB binding pockets, and all-atom molecular dynamics simulations in different subregions of the OM model. Using these descriptors and the measured inhibitory concentrations, we design a statistical protocol to identify predictors of OM permeation/inhibition. We find consistent rules across most of our data highlighting the role of the interaction between the compounds and the OM. An implementation of the rules uncovered in our study is shown, and it demonstrates the accuracy of our approach in a set of previously unseen compounds. Our analysis sheds new light on the key properties drug candidates need to effectively permeate/inhibit P. aeruginosa, and opens the gate to similar data-driven studies in other Gram-negative pathogens.