2024-10-30 ワシントン大学セントルイス校
ワシントン大学医学部の研究チームは、腫瘍の内部構造とその複雑さを3D技術で詳細にマッピングすることに成功しました。この技術は、腫瘍内の細胞やその周囲の環境がどのように組織されているかを立体的に捉えることができ、従来の2Dスライスでは捉えきれなかった腫瘍の立体的な構造や細胞間の相互作用を明確にすることが可能です。研究者たちは、さまざまな種類の腫瘍について、異なる細胞がどのように配置され、相互作用しているかを解析し、腫瘍の成長や進行に寄与する要因を特定することを目指しています。
<関連情報>
- https://source.washu.edu/2024/10/complexity-of-tumors-revealed-in-3d/
- https://www.nature.com/articles/s41586-024-08087-4
- https://www.nature.com/articles/s43018-024-00773-6
- https://www.nature.com/articles/s41592-024-02438-9
2Dおよび3D空間における腫瘍の進化と微小環境の相互作用 Tumour evolution and microenvironment interactions in 2D and 3D space
Chia-Kuei Mo,Jingxian Liu,Siqi Chen,Erik Storrs,Andre Luiz N. Targino da Costa,Andrew Houston,Michael C. Wendl,Reyka G. Jayasinghe,Michael D. Iglesia,Cong Ma,John M. Herndon,Austin N. Southard-Smith,Xinhao Liu,Jacqueline Mudd,Alla Karpova,Andrew Shinkle,S. Peter Goedegebuure,Abdurrahman Taha Mousa Ali Abdelzaher,Peng Bo,Lauren Fulghum,Samantha Livingston,Metin Balaban,Angela Hill,Joseph E. Ippolito,… Li Ding
Nature Published:30 October 2024
DOI:https://doi.org/10.1038/s41586-024-08087-4
Abstract
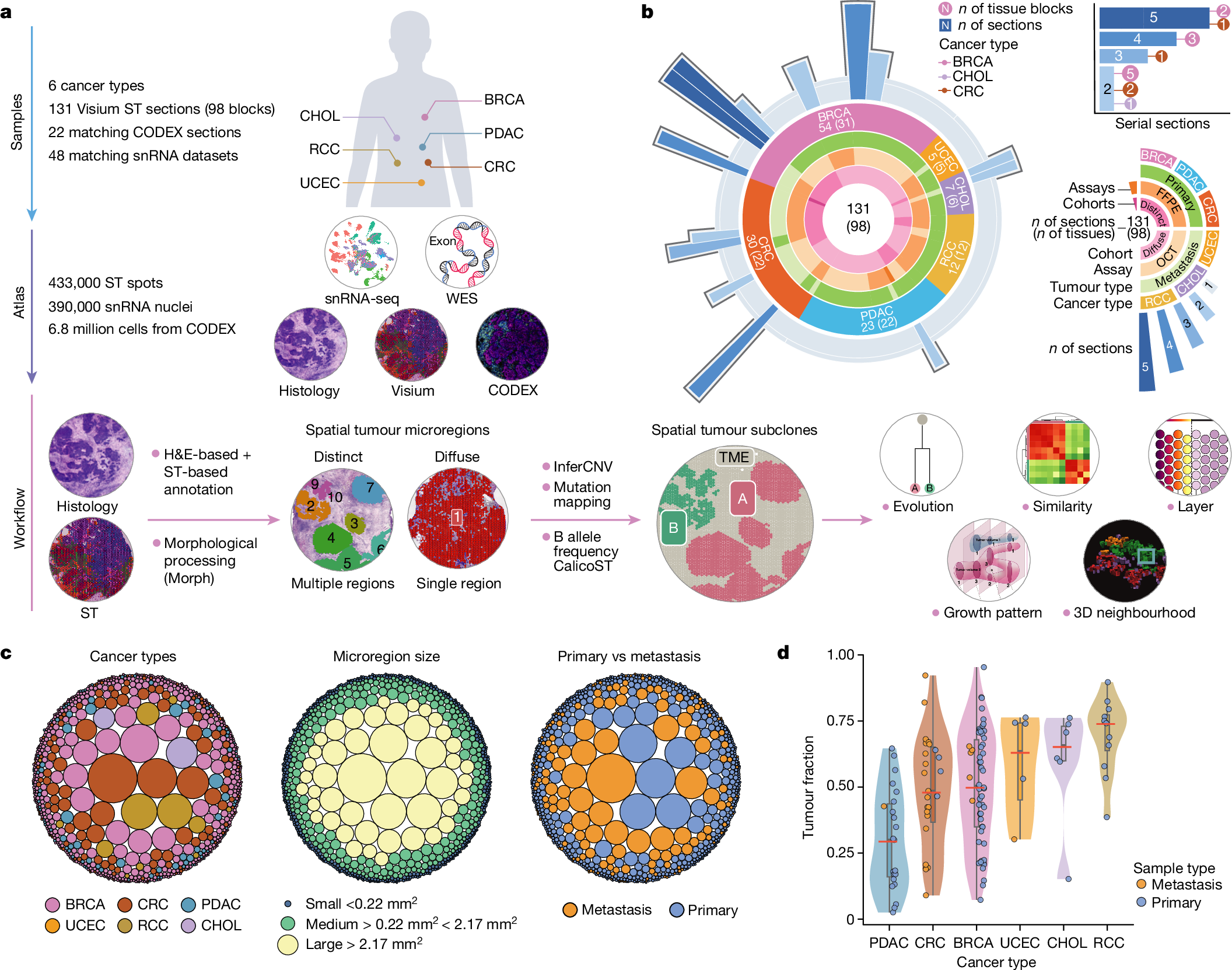
To study the spatial interactions among cancer and non-cancer cells1, we here examined a cohort of 131 tumour sections from 78 cases across 6 cancer types by Visium spatial transcriptomics (ST). This was combined with 48 matched single-nucleus RNA sequencing samples and 22 matched co-detection by indexing (CODEX) samples. To describe tumour structures and habitats, we defined ‘tumour microregions’ as spatially distinct cancer cell clusters separated by stromal components. They varied in size and density among cancer types, with the largest microregions observed in metastatic samples. We further grouped microregions with shared genetic alterations into ‘spatial subclones’. Thirty five tumour sections exhibited subclonal structures. Spatial subclones with distinct copy number variations and mutations displayed differential oncogenic activities. We identified increased metabolic activity at the centre and increased antigen presentation along the leading edges of microregions. We also observed variable T cell infiltrations within microregions and macrophages predominantly residing at tumour boundaries. We reconstructed 3D tumour structures by co-registering 48 serial ST sections from 16 samples, which provided insights into the spatial organization and heterogeneity of tumours. Additionally, using an unsupervised deep-learning algorithm and integrating ST and CODEX data, we identified both immune hot and cold neighbourhoods and enhanced immune exhaustion markers surrounding the 3D subclones. These findings contribute to the understanding of spatial tumour evolution through interactions with the local microenvironment in 2D and 3D space, providing valuable insights into tumour biology.
クロマチンアクセシビリティと転写動態の違いが乳がんのサブタイプとその系譜を規定する Differential chromatin accessibility and transcriptional dynamics define breast cancer subtypes and their lineages
Michael D. Iglesia,Reyka G. Jayasinghe,Siqi Chen,Nadezhda V. Terekhanova,John M. Herndon,Erik Storrs,Alla Karpova,Daniel Cui Zhou,Nataly Naser Al Deen,Andrew T. Shinkle,Rita Jui-Hsien Lu,Wagma Caravan,Andrew Houston,Yanyan Zhao,Kazuhito Sato,Preet Lal,Cherease Street,Fernanda Martins Rodrigues,Austin N. Southard-Smith,André Luiz N. Targino da Costa,Houxiang Zhu,Chia-Kuei Mo,Lisa Crowson,Robert S. Fulton,… Li Ding
Nature Cancer Published:30 October 2024
DOI:https://doi.org/10.1038/s43018-024-00773-6
Abstract
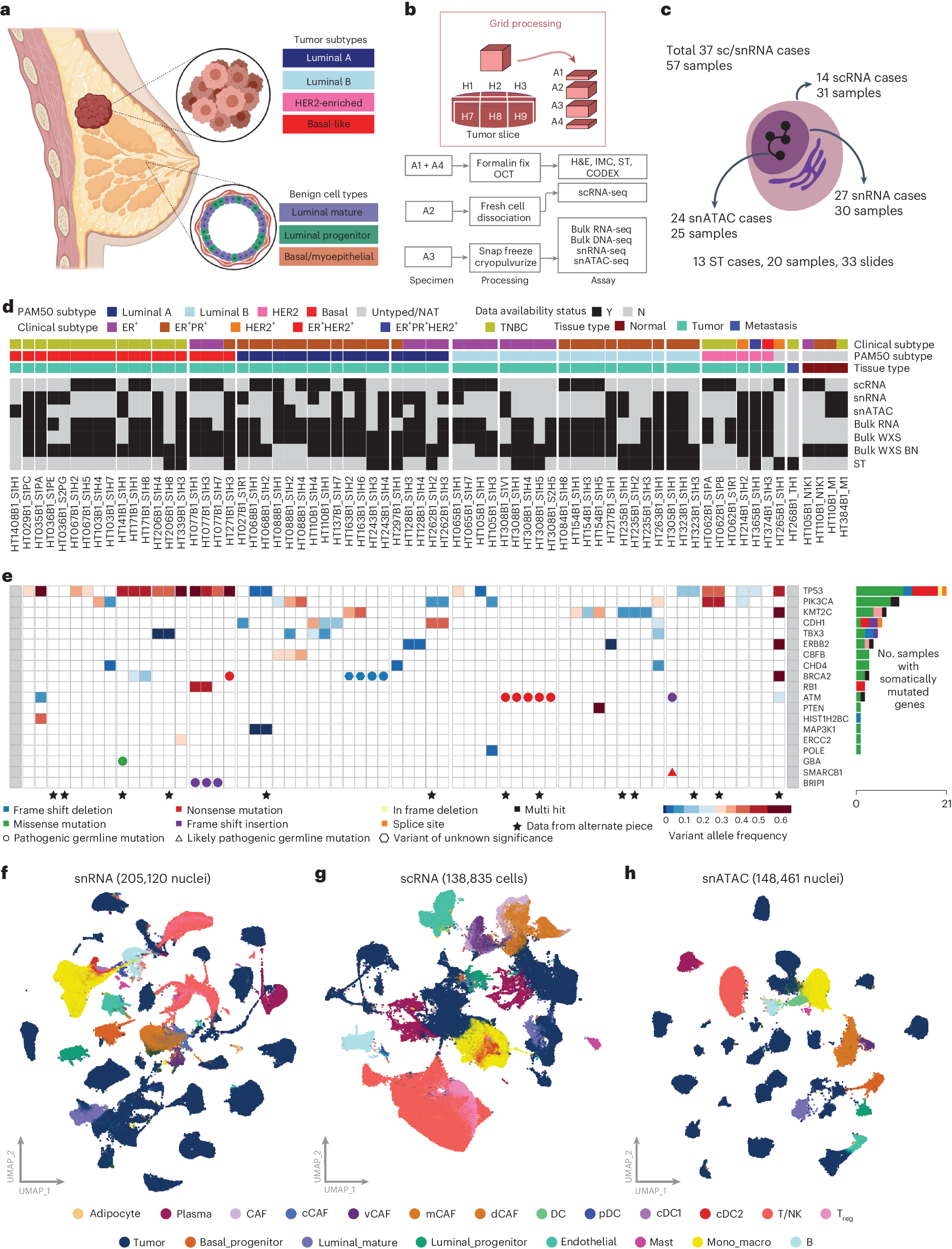
Breast cancer (BC) is defined by distinct molecular subtypes with different cells of origin. The transcriptional networks that characterize the subtype-specific tumor-normal lineages are not established. In this work, we applied bulk, single-cell and single-nucleus multi-omic techniques as well as spatial transcriptomics and multiplex imaging on 61 samples from 37 patients with BC to show characteristic links in gene expression and chromatin accessibility between BC subtypes and their putative cells of origin. Regulatory network analysis of transcription factors underscored the importance of BHLHE40 in luminal BC and luminal mature cells and KLF5 in basal-like tumors and luminal progenitor cells. Furthermore, we identify key genes defining the basal-like (SOX6 and KCNQ3) and luminal A/B (FAM155A and LRP1B) lineages. Exhausted CTLA4-expressing CD8+ T cells were enriched in basal-like BC, suggesting an altered means of immune dysfunction. These findings demonstrate analysis of paired transcription and chromatin accessibility at the single-cell level is a powerful tool for investigating cancer lineage and highlight transcriptional networks that define basal and luminal BC lineages.
空間分解トランスクリプトミクスから対立遺伝子特異的コピー数異常と腫瘍系統地理を推定する Inferring allele-specific copy number aberrations and tumor phylogeography from spatially resolved transcriptomics
Cong Ma,Metin Balaban,Jingxian Liu,Siqi Chen,Michael J. Wilson,Christopher H. Sun,Li Ding & Benjamin J. Raphael
Nature Methods Published:30 October 2024
DOI:https://doi.org/10.1038/s41592-024-02438-9
Abstract
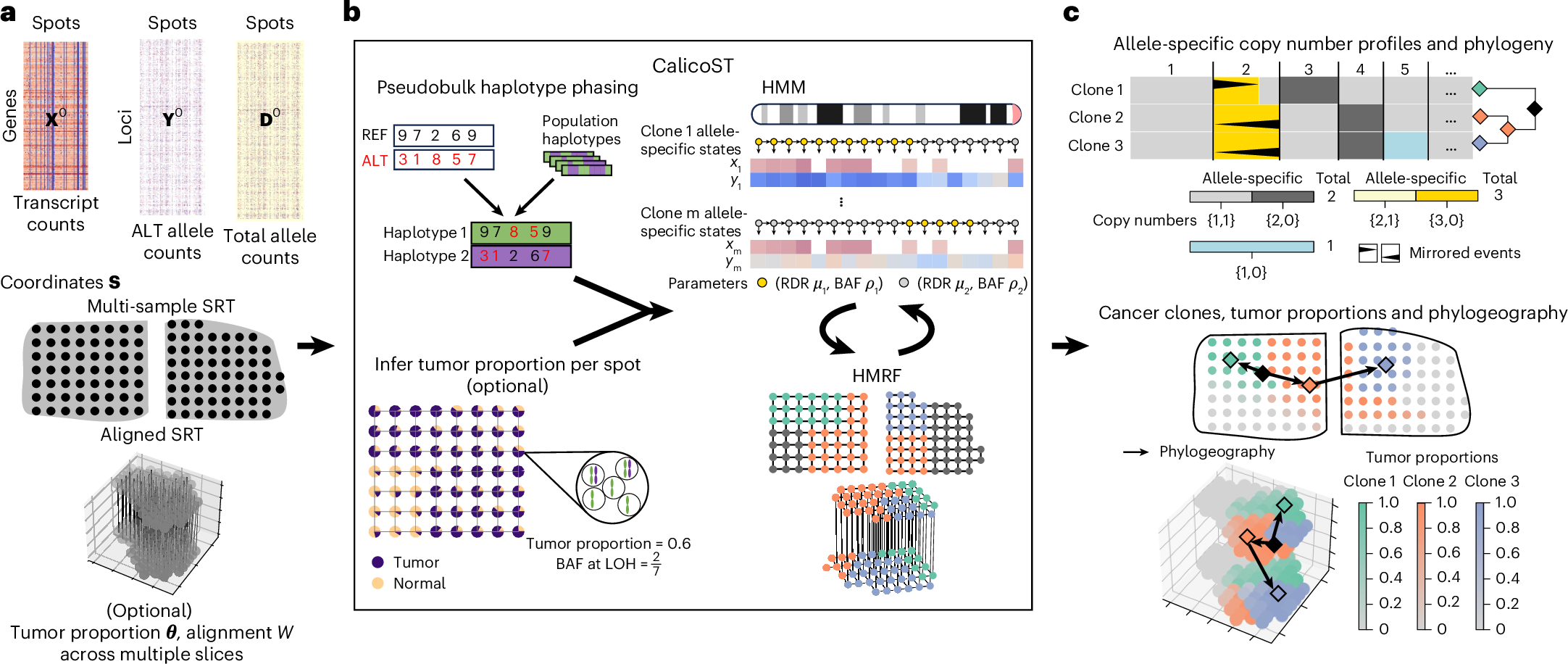
Analyzing somatic evolution within a tumor over time and across space is a key challenge in cancer research. Spatially resolved transcriptomics (SRT) measures gene expression at thousands of spatial locations in a tumor, but does not directly reveal genomic aberrations. We introduce CalicoST, an algorithm to simultaneously infer allele-specific copy number aberrations (CNAs) and reconstruct spatial tumor evolution, or phylogeography, from SRT data. CalicoST identifies important classes of CNAs—including copy-neutral loss of heterozygosity and mirrored subclonal CNAs—that are invisible to total copy number analysis. Using nine patients’ data from the Human Tumor Atlas Network, CalicoST achieves an average accuracy of 86%, approximately 21% higher than existing methods. CalicoST reconstructs a tumor phylogeography in three-dimensional space for two patients with multiple adjacent slices. CalicoST analysis of multiple SRT slices from a cancerous prostate organ reveals mirrored subclonal CNAs on the two sides of the prostate, forming a bifurcating phylogeography in both genetic and physical space.