2024-11-08 ロスアラモス国立研究所(LANL)
<関連情報>
- https://discover.lanl.gov/news/1107-ai-genomics/
- https://academic.oup.com/nar/article/52/19/e91/7756581?login=false
DNA呼吸とディープラーニング基礎モデルの統合により、ヒト転写因子のゲノムワイド結合予測が進歩 DNA breathing integration with deep learning foundational model advances genome-wide binding prediction of human transcription factors
Anowarul Kabir, Manish Bhattarai, Selma Peterson, Yonatan Najman-Licht, Kim Ø Rasmussen, Amarda Shehu, Alan R Bishop, Boian Alexandrov, Anny Usheva
Nucleic Acids Research Published:13 September 2024
DOI:https://doi.org/10.1093/nar/gkae783
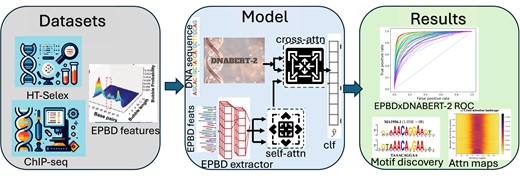
Graphical Abstract
Abstract
It was previously shown that DNA breathing, thermodynamic stability, as well as transcriptional activity and transcription factor (TF) bindings are functionally correlated. To ascertain the precise relationship between TF binding and DNA breathing, we developed the multi-modal deep learning model EPBDxDNABERT-2, which is based on the Extended Peyrard-Bishop-Dauxois (EPBD) nonlinear DNA dynamics model. To train our EPBDxDNABERT-2, we used chromatin immunoprecipitation sequencing (ChIP-Seq) data comprising 690 ChIP-seq experimental results encompassing 161 distinct TFs and 91 human cell types. EPBDxDNABERT-2 significantly improves the prediction of over 660 TF-DNA, with an increase in the area under the receiver operating characteristic (AUROC) metric of up to 9.6% when compared to the baseline model that does not leverage DNA biophysical properties. We expanded our analysis to in vitro high-throughput Systematic Evolution of Ligands by Exponential enrichment (HT-SELEX) dataset of 215 TFs from 27 families, comparing EPBD with established frameworks. The integration of the DNA breathing features with DNABERT-2 foundational model, greatly enhanced TF-binding predictions. Notably, EPBDxDNABERT-2, trained on a large-scale multi-species genomes, with a cross-attention mechanism, improved predictive power shedding light on the mechanisms underlying disease-related non-coding variants discovered in genome-wide association studies.


