2025-06-12 国立遺伝学研究所
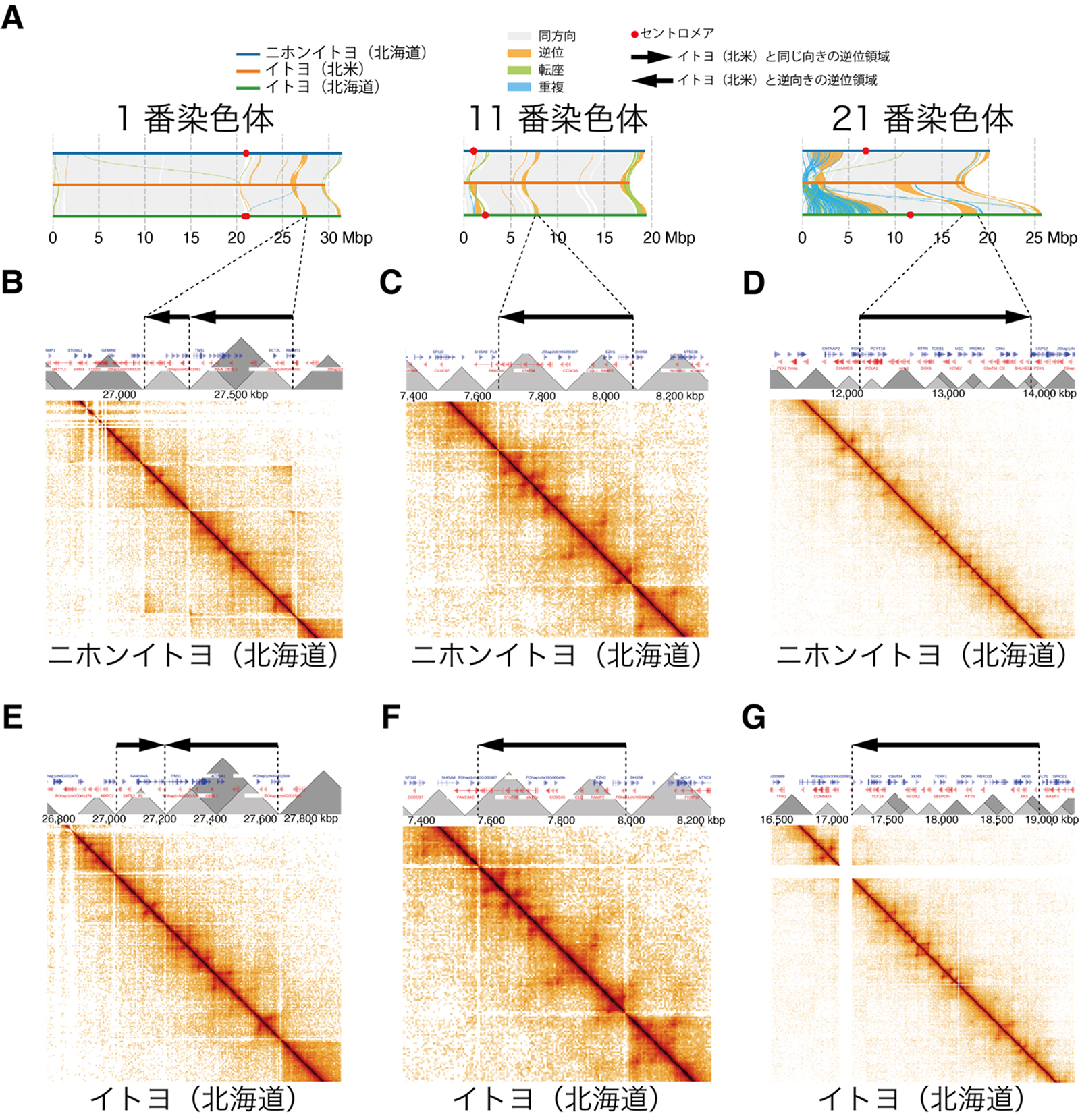
図:逆位領域の切断点(末端)とTAD(グレーの三角形)の境界の頻繁な一致を示す図.(A)ニホンイトヨ(北海道),イトヨ(北米),イトヨ(北海道)を比較した場合に見つかった代表的な4つの逆位を示す.ニホンイトヨ(北海道)(B-D),およびイトヨ(北海道)(E-G)におけるTAD構造と逆位の位置関係.ヒートマップはHi-C法により検出された,DNA領域間の物理的な接触頻度を示す.接触頻度が高い領域がTADとして推定される.
<関連情報>
- https://www.nig.ac.jp/nig/ja/2025/06/research-highlights_ja/rh20250531.html
- https://onlinelibrary.wiley.com/doi/10.1111/mec.17814
3次元ゲノムの解析により、トゲウオの遺伝子流動の障壁となる逆行性のブレイクポイントが明らかになった 3D Genome Constrains Breakpoints of Inversions That Can Act as Barriers to Gene Flow in the Stickleback
Yo Y. Yamasaki, Atsushi Toyoda, Mitsutaka Kadota, Shigehiro Kuraku, Jun Kitano
Molecular Ecology Published: 31 May 2025
DOI:https://doi.org/10.1111/mec.17814
ABSTRACT
DNA within the nucleus is organised into a well-regulated three-dimensional (3D) structure. However, how such 3D genome structures influence speciation processes remains largely elusive. Recent studies have shown that 3D genome structures influence mutation rates, including the occurrence of chromosomal rearrangement. For example, breakpoints of chromosomal rearrangements tend to be located at topologically associating domain (TAD) boundaries. Here, we hypothesised that TAD structures may constrain the location of chromosomal inversions and thereby shape the genomic landscape of divergence between species with ongoing gene flow, given that inversions can act as barriers to gene flow. To test this hypothesis, we used a pair of Japanese stickleback species, Gasterosteus nipponicus (Japan Sea stickleback) and G. aculeatus (three-spined stickleback). We first constructed chromosome-scale genome assemblies of both species using high fidelity long reads and high-resolution proximity ligation data and identified several chromosomal inversions. Second, via population genomic analyses, we revealed higher genetic differentiation in inverted regions than in colinear regions and no gene flow within inversions, which contrasts with the significant gene flow in colinear regions. Third, using Hi-C data, we revealed 3D genome structures of sticklebacks, delineated by A/B compartments and TADs. Finally, we found that inversion breakpoints tend to be located at TAD boundaries. Thus, our study demonstrates that the 3D genome constrains breakpoints of inversions that can act as barriers to gene flow in the stickleback. Further integration of 3D genome analyses with population genomics could provide novel insights into how the 3D genome influences speciation.