2025-07-07 バッファロー大学 (UB)
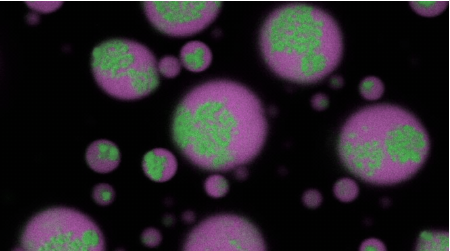
RNA clusters (green colored) gradually disassemble within biomolecular condensates (magenta colored), returning the RNA to a soluble state. This occured after an antisense oligonucleotide engineered by the team of Priya Banerjee was introduced to the condensate.
<関連情報>
- https://www.buffalo.edu/news/releases/2025/07/disassemble-RNA-clusters-linked-to-neurological-diseases.html
- https://www.nature.com/articles/s41557-025-01847-3
多成分からなる生体分子凝縮体のコア内部における液体から固体への転移に伴うホモ型RNAクラスター形成 Homotypic RNA clustering accompanies a liquid-to-solid transition inside the core of multi-component biomolecular condensates
Tharun Selvam Mahendran,Gable M. Wadsworth,Anurag Singh,Ritika Gupta & Priya R. Banerjee
Nature Chemistry Published:02 July 2025
DOI:https://doi.org/10.1038/s41557-025-01847-3
Abstract
RNA-driven condensation plays a central role in organizing and regulating ribonucleoprotein granules within cells. Disruptions to this process—such as the aberrant aggregation of repeat-expanded RNA—are associated with numerous neurological disorders. Here we study the role of biomolecular condensates in irreversible RNA aggregation. We find that physiologically relevant and disease-associated repeat RNAs spontaneously undergo an age-dependent percolation transition inside multi-component condensates to form nanoscale clusters. Homotypic RNA clusters drive the emergence of multi-phasic condensate structures, with an RNA-rich solid core surrounded by an RNA-depleted fluid shell. The timescale of RNA clustering is determined by sequence, secondary structure and repeat length. Importantly, G3BP1, the core scaffold of stress granules, introduces heterotypic buffering to homotypic RNA–RNA interactions and prevents RNA clustering in an ATP-independent manner. Our work suggests that biomolecular condensates can act as sites for RNA aggregation and highlights the chaperone-like function of RNA-binding proteins against aberrant RNA phase transitions.

