2025-07-14 ワシントン大学セントルイス校
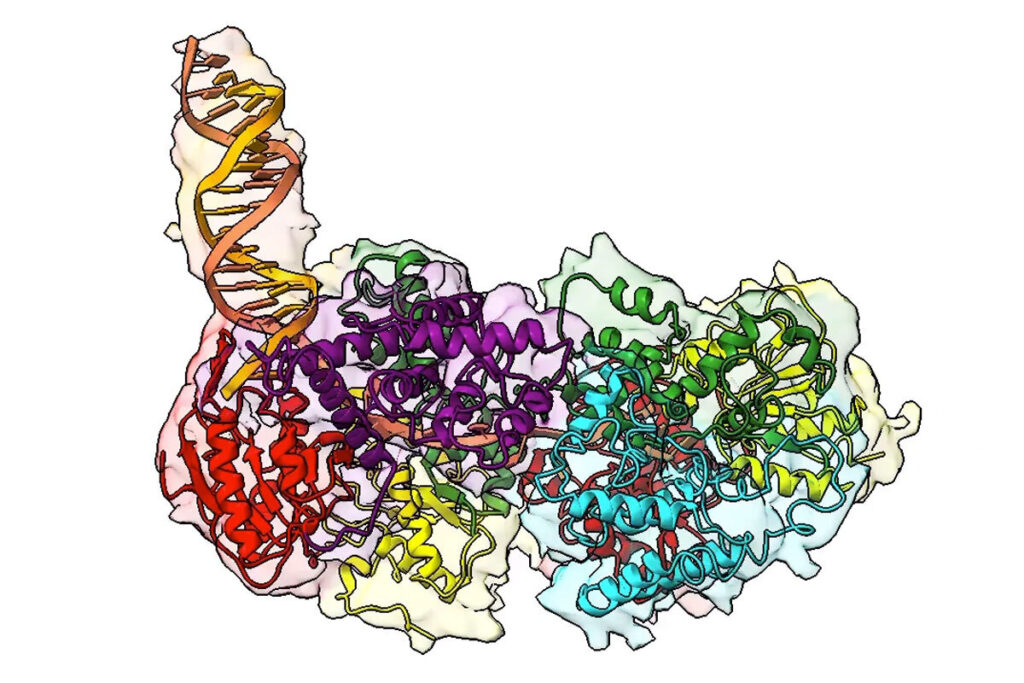
A still image from a three-dimensional video rendering shows a key protein involved in DNA repair in many organisms. Here, the protein is bound to a DNA molecule (the protruding orange and yellow double helix) and beginning to “unzip” its two strands to allow for DNA repair. (Image: Galburt Lab/WashU Medicine)
<関連情報>
- https://source.washu.edu/2025/07/key-component-to-cell-division-unveiled-in-3d/
- https://www.pnas.org/doi/10.1073/pnas.2422330122
UvrDファミリーヘリカーゼの二量化と活性化の構造的基盤 Structural basis for dimerization and activation of UvrD-family helicases
Ankita Chadda, Binh Nguyen, Timothy M. Lohman, and Eric A. Galburt
Proceedings of the National Academy of Sciences Published:March 6, 2025
DOI:https://doi.org/10.1073/pnas.2422330122
Significance
The integrity and flow of genetic material depend on enzymes for the maintenance and transfer of the contained information. More specifically, DNA helicases are involved in DNA replication, transcription, homologous recombination, and DNA repair. UvrD-family helicases are found from bacteria to man and physical biochemistry has shown that these enzymes must dimerize to become active helicases in the absence of other factors. However, only monomeric molecular structures have been observed. Here, we reveal the structural mechanism of helicase activation through dimerization by elucidating dimeric structures of Mycobacterium tuberculosis UvrD1. Our results support the biochemistry, argue against previous interpretations of the monomeric structures, and coalesce the field into a consistent model of how this class of enzymes function.
Abstract
UvrD-family helicases are superfamily 1A motor proteins that function during DNA replication, recombination, repair, and transcription. UvrD family monomers translocate along single-stranded (ss) DNA but need to be activated by dimerization to unwind DNA in the absence of force or accessory factors. However, prior structural studies have only revealed monomeric complexes. Here, we report the first structures of a dimeric UvrD-family helicase, Mycobacterium tuberculosis UvrD1, both free and bound to a DNA junction. In each structure, the dimer interface occurs between the 2B subdomains of each subunit. The apo UvrD1 dimer is observed in symmetric compact and extended forms indicating substantial flexibility. This symmetry is broken in the DNA-bound dimer complex with leading and trailing subunits adopting distinct conformations. Biochemical experiments reveal that the Escherichia coli UvrD dimer shares the same 2B–2B interface. In contrast to the dimeric structures, an inactive, autoinhibited UvrD1 DNA-bound monomer structure reveals 2B subdomain–DNA contacts that are likely inhibitory. The major reorientation of the 2B subdomains that occurs upon UvrD1 dimerization prevents these duplex DNA interactions, thus relieving the autoinhibition. These structures reveal that the 2B subdomain serves a major regulatory role rather than participating directly in DNA unwinding.