2025-09-24 ワシントン州立大学(WSU)
<関連情報>
- https://news.wsu.edu/press-release/2025/09/24/key-to-the-riddle-of-sleep-may-be-linked-to-bacteria/
- https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2025.1608302/full
- https://www.sciencedirect.com/science/article/pii/S108707922500098X
細菌ペプチドグリカン濃度は脳領域、時間帯、睡眠不足によって変動する Bacterial peptidoglycan levels have brain area, time of day, and sleep loss-induced fluctuations
Erika L. English,James M. Krueger
Frontiers in Neuroscience Published:16 July 2025
DOI:https://doi.org/10.3389/fnins.2025.1608302
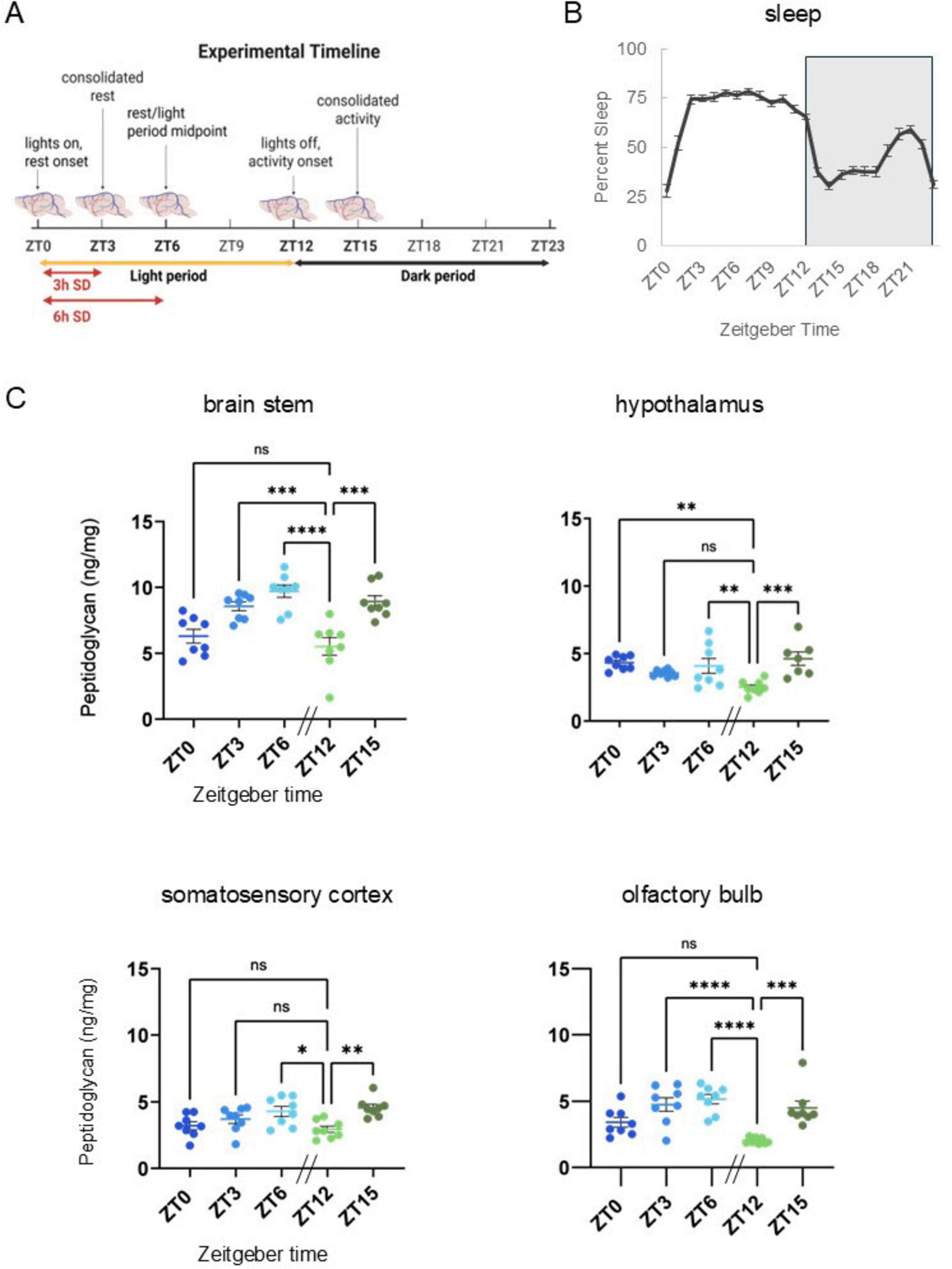
Sleep-inducing bacterial cell wall components isolated from brain and urine of sleep deprived animals were identified as peptidoglycan (PG) and muropeptides in the 1980s. Following host detection of PG/muropeptides, downstream signaling mechanisms include release of effector molecules, e.g., cytokines involved in sleep regulation. Understanding of physiological brain PG changes has remained limited, in part due to the historic difficulties of PG quantitation. Herein, we report murine brain PG levels in multiple brain areas within the context of animals’ rest-wake cycles and after sleep loss. Significant time-of-day changes in brain PG levels occurred in all brain areas; lowest levels occurred during the transition from rest to wake periods, at zeitgeber time 12 (ZT12). Highest levels of PG were in brainstem while olfactory bulb, hypothalamic, and cortical PG levels were lower. After 3 h of sleep disruption, PG levels increased in the somatosensory cortex, but decreased in brainstem, and hypothalamus. After 6 h of sleep disruption, PG increased in the brainstem and olfactory bulb compared to control levels. Further, RNA-seq analyses of somatosensory cortical tissue was used to assess sleep loss-dependent changes in genes previously linked to PG. Multiple PG-related genes had altered expression with sleep loss including PG binding and signaling molecules, e.g., Pglyrp1 and Nfil3. In summary, brain PG levels were dependent on time of day, brain area, and sleep history. Further, sleep loss altered brain gene expression for PG-linked genes. Collectively, these data are consistent with the hypothesis that microbe-host symbiotic interactions are involved in murine sleep regulatory mechanisms.
局所的およびニッチに適応した睡眠調節機構がホロバイオントの状態を包含する Local and niche-adapted sleep regulatory mechanisms encompass the holobiont condition
Erika L. English, James M. Krueger
Sleep Medicine Reviews Available online: 5 August 2025
DOI:https://doi.org/10.1016/j.smrv.2025.102145
Abstract
We posit that organism sleep is regulated from the interactions between two semi-autonomous regulatory systems, the classic sleep/wake regulatory circuits and a local cell activity-driven system. Sleep regulatory circuits mold local sleep into a species’ niche but are not required for sleep. In contrast, local sleep mechanisms initiate sleep-like states in small networks and are responsive to microbial pattern recognition receptors. Local sleep-like phenomena manifest in brain in vivo and in vitro. Sleep regulatory substances e.g., cytokines and adenosine, are released by cell activity and as such provide an index of neural network activity. They are evolutionary ancient molecules existing prior to the evolution of complex vertebrate sleep. Further, bacterial cell wall components can initiate sleep in insomniac mammals lacking key hypothalamic regulatory circuits, suggesting our ancient holobiont condition underlies sleep regulation. We review how interleukin-1 promotes sleep/sleep-like states at various tissue organization levels. We provide a basis for understanding sleep as an emergent property of cellular networks and a process beginning at the cellular level and progressing as modified by multiple physiological regulatory circuits to whole animal sleep. We conclude; sleep mechanisms are shared across various levels of tissue organization and are part of interspecies regulatory networks.


