2025-09-30 筑波大学
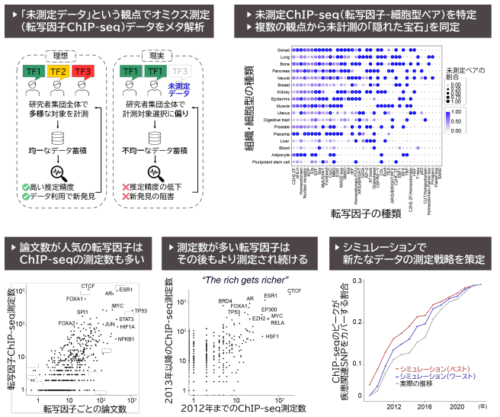
図 本研究で⾏った未測定転写因⼦-組織・細胞型ペアの抽出と戦略的活⽤の概要
<関連情報>
- https://www.tsukuba.ac.jp/journal/medicine-health/20250930140000.html
- https://www.tsukuba.ac.jp/journal/pdf/p20250930140000.pdf
- https://academic.oup.com/bfg/article/doi/10.1093/bfgp/elaf016/8266060
未測定のヒト転写因子ChIP-seqデータが機能ゲノム学に与える影響と戦略的優先順位付けの必要性 Unmeasured human transcription factor ChIP-seq data shape functional genomics and demand strategic prioritization
Saeko Tahara, Haruka Ozaki
Briefings in Functional Genomics Published::30 September 2025
DOI:https://doi.org/10.1093/bfgp/elaf016
Abstract
Transcription factor (TF) chromatin immunoprecipitation followed by sequencing (ChIP-seq) is essential for identifying genome-wide TF-binding sites (TFBSs), and the collected datasets offer a variety of opportunities for downstream analyses such as inference of gene regulatory network and prediction for effects of single-nucleotide polymorphisms (SNPs) on TFBSs. Although TF ChIP-seq data continue to accumulate in public databases, comprehensive coverage of biologically relevant TF-sample pairs (i.e. combination of targeted TF and cell type) remains elusive. This is due to the need for TF-specific antibodies and large cell numbers, limiting feasible TF–cell type combinations. Moreover, ChIP-seq is measurable when the TF is expressed in the target cell type. Thus, defining the full space of biologically relevant TF–sample pairs—including both measured and unmeasured—is essential to assess and improve dataset comprehensiveness. Here, we investigated publicly available human TF ChIP-seq datasets and introduced the concept of unmeasured TF-sample pairs, defined as biologically relevant TF–sample combinations for which ChIP-seq experiments have not yet been performed. Notably, many expressed TFs in specific cell types remain unmeasured by ChIP-seq, affecting the coverage of regulatory regions revealed by TF ChIP-seq and genome-wide association study–SNP analyses. Furthermore, we propose practical strategies to efficiently supplement currently unmeasured data and discuss how these approaches can significantly enhance data-driven research. The database of unmeasured human TF–sample pairs is publicly accessible at https://moccs-db.shinyapps.io/Unmeasured_shiny_v1/, facilitating the systematic expansion of TF ChIP-seq datasets and thereby enhancing our comprehension of gene regulatory mechanisms.