2025-11-21 国立成育医療研究センター
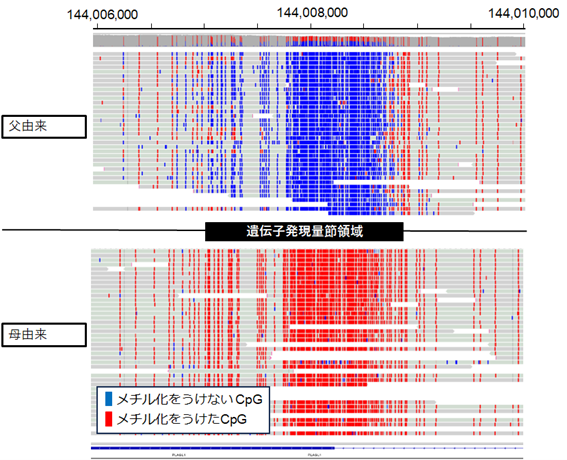
【図1.LRSによるDMRのメチル化解析結果】
<関連情報>
- https://www.ncchd.go.jp/press/2025/1121.html
- https://www.ncchd.go.jp/press/2025/1121.pdf
- https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-025-01559-w
差次的メチル化領域およびインプリンティング障害関連遺伝子におけるDNAメチル化を評価するための包括的なロングリードシーケンシングシステム A comprehensive long-read sequencing system to assess DNA methylation at differentially methylated regions and imprinting-disorder-related genes
Tatsuki Urakawa,Atsushi Hattori,Yasuko Ogiwara,Hayate Masubuchi,Mizuho Igarashi,Sayuri Nakamura,Kaori Hara-Isono,Keisuke Ishiwata,Hiroko Ogata-Kawata,Hiromi Kamura,Yoko Kuroki,Kazuhiko Nakabayashi,Maki Fukami & Masayo Kagami
Genome Medicine Published:18 November 2025
DOI:https://doi.org/10.1186/s13073-025-01559-w
Abstract
Background
Imprinted genes are expressed in a parental-origin–specific manner. The imprinted regions including imprinted genes have differentially methylated regions (DMRs) with different 5-methylcytosine (5mC) patterns for CpGs on each parental allele, and DMRs function as imprinting control centers. Aberrant expression of the imprinted genes caused by structural variants involving DMRs, single-nucleotide variants in imprinted genes, uniparental disomy, and epimutation lead to imprinting disorders (IDs). Nanopore-based targeted long-read sequencing (T-LRS) can obtain sequence reads 10–100 kb long together with information on DNA methylation in each CpG and is cost-effective compared to whole-genome LRS. T-LRS is a valuable tool for efficient genetic testing for IDs and has great potential to elucidate the regulatory mechanisms in the imprinted regions. However, there is no T-LRS system targeting all ID-related regions.
Methods
We conducted T-LRS targeting 78 DMRs and 22 genes in peripheral blood leukocytes from six healthy controls and set the normal range of methylation index (MI) for each CpG within the DMRs. To clarify the properties of DMRs, we compared MIs in CpGs within DMRs between haplotypes in 78 DMRs. To evaluate the usefulness of T-LRS, we conducted T-LRS on two previously reported patients with multi-locus imprinting disturbance (MLID) having pathogenic variants in MLID-causative genes and compared the MIs in CpGs within DMRs with those measured by array-based methylation analysis.
Results
The median number of reads with 5mC and unmethylated cytosine in all DMRs in the six controls was over 40. We defined the normal range of MI for all CpGs in each allele and the total, and classified 78 DMRs into three categories, namely, 33 Complete-DMRs, 25 Partial-DMRs, and 20 Non-DMRs, based on the average of six controls for the median of differences of MIs in CpGs between haplotypes. We confirmed by T-LRS pathogenic variants in MLID-causative genes in patients with MLID. The patients’ methylation defect patterns in T-LRS were similar to those in array-based methylation analysis, although T-LRS showed additional aberrantly methylated DMRs.
Conclusions
We established a T-LRS system targeting all ID-related regions, defined standard MI ranges in CpGs on each parental allele, and demonstrated the usefulness of T-LRS.